The repertoire of G protein-coupled receptors in the sea squirt Ciona intestinalis
- PMID: 18452600
- PMCID: PMC2396169
- DOI: 10.1186/1471-2148-8-129
The repertoire of G protein-coupled receptors in the sea squirt Ciona intestinalis
Abstract
Background: G protein-coupled receptors (GPCRs) constitute a large family of integral transmembrane receptor proteins that play a central role in signal transduction in eukaryotes. The genome of the protochordate Ciona intestinalis has a compact size with an ancestral complement of many diversified gene families of vertebrates and is a good model system for studying protochordate to vertebrate diversification. An analysis of the Ciona repertoire of GPCRs from a comparative genomic perspective provides insight into the evolutionary origins of the GPCR signalling system in vertebrates.
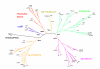
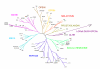
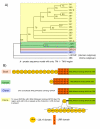
Results: We have identified 169 gene products in the Ciona genome that code for putative GPCRs. Phylogenetic analyses reveal that Ciona GPCRs have homologous representatives from the five major GRAFS (Glutamate, Rhodopsin, Adhesion, Frizzled and Secretin) families concomitant with other vertebrate GPCR repertoires. Nearly 39% of Ciona GPCRs have unambiguous orthologs of vertebrate GPCR families, as defined for the human, mouse, puffer fish and chicken genomes. The Rhodopsin family accounts for ~68% of the Ciona GPCR repertoire wherein the LGR-like subfamily exhibits a lineage specific gene expansion of a group of receptors that possess a novel domain organisation hitherto unobserved in metazoan genomes.
Conclusion: Comparison of GPCRs in Ciona to that in human reveals a high level of orthology of a protochordate repertoire with that of vertebrate GPCRs. Our studies suggest that the ascidians contain the basic ancestral complement of vertebrate GPCR genes. This is evident at the subfamily level comparisons since Ciona GPCR sequences are significantly analogous to vertebrate GPCR subfamilies even while exhibiting Ciona specific genes. Our analysis provides a framework to perform future experimental and comparative studies to understand the roles of the ancestral chordate versions of GPCRs that predated the divergence of the urochordates and the vertebrates.
Figures
Similar articles
-
Remarkable similarities between the hemichordate (Saccoglossus kowalevskii) and vertebrate GPCR repertoire.Gene. 2013 Sep 10;526(2):122-33. doi: 10.1016/j.gene.2013.05.005. Epub 2013 May 15. Gene. 2013. PMID: 23685280
-
The GPCR repertoire in the demosponge Amphimedon queenslandica: insights into the GPCR system at the early divergence of animals.BMC Evol Biol. 2014 Dec 21;14:270. doi: 10.1186/s12862-014-0270-4. BMC Evol Biol. 2014. PMID: 25528161 Free PMC article.
-
The G protein-coupled receptors in the pufferfish Takifugu rubripes.BMC Bioinformatics. 2011 Feb 15;12 Suppl 1(Suppl 1):S3. doi: 10.1186/1471-2105-12-S1-S3. BMC Bioinformatics. 2011. PMID: 21342560 Free PMC article.
-
Omics Studies for the Identification of Ascidian Peptides, Cognate Receptors, and Their Relevant Roles in Ovarian Follicular Development.Front Endocrinol (Lausanne). 2022 Mar 7;13:858885. doi: 10.3389/fendo.2022.858885. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35321341 Free PMC article. Review.
-
Superfamily of G-protein coupled receptors (GPCRs)--extraordinary and outstanding success of evolution.Postepy Hig Med Dosw (Online). 2014 Oct 31;68:1225-37. doi: 10.5604/17322693.1127326. Postepy Hig Med Dosw (Online). 2014. PMID: 25380205 Review.
Cited by
-
Molecular cloning and expression analysis of mc5r like genes (mc5rl) in Ruditapes philippinarum (Manila clam) after aerial exposure and low-temperature stress.Mol Biol Rep. 2020 Nov;47(11):8891-8901. doi: 10.1007/s11033-020-05941-2. Epub 2020 Oct 31. Mol Biol Rep. 2020. PMID: 33128687
-
Constitutively active glucagon receptor drives high blood glucose in birds.Nature. 2025 May;641(8065):1287-1297. doi: 10.1038/s41586-025-08811-8. Epub 2025 Mar 3. Nature. 2025. PMID: 40031956 Free PMC article.
-
Immune diversity in lophotrochozoans, with a focus on recognition and effector systems.Comput Struct Biotechnol J. 2023 Mar 22;21:2262-2275. doi: 10.1016/j.csbj.2023.03.031. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37035545 Free PMC article. Review.
-
The pre-vertebrate origins of neurogenic placodes.Nature. 2015 Aug 27;524(7566):462-5. doi: 10.1038/nature14657. Epub 2015 Aug 10. Nature. 2015. PMID: 26258298 Free PMC article.
-
Serotonin Receptors and Their Involvement in Melanization of Sensory Cells in Ciona intestinalis.Cells. 2023 Apr 13;12(8):1150. doi: 10.3390/cells12081150. Cells. 2023. PMID: 37190059 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

