Ca2+-dimethylphosphate complex formation: providing insight into Ca2+-mediated local dehydration and membrane fusion in cells
- PMID: 18452809
- PMCID: PMC4580261
- DOI: 10.1016/j.cellbi.2008.03.002
Ca2+-dimethylphosphate complex formation: providing insight into Ca2+-mediated local dehydration and membrane fusion in cells
Abstract
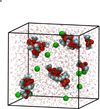
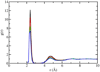
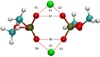
Earlier studies using X-ray diffraction, light scattering, photon correlation spectroscopy, and atomic force microscopy, strongly suggest that SNARE-induced membrane fusion in cells proceeds as a result of calcium bridging opposing bilayers. The bridging of phospholipid heads groups in the opposing bilayers by calcium leads to the release of water from hydrated Ca(2+) ions as well as the loosely coordinated water at PO-lipid head groups. Local dehydration of phospholipid head groups and the calcium, bridging opposing bilayers, then leads to destabilization of the lipid bilayers and membrane fusion. This hypothesis was tested in the current study by atomistic molecular dynamic simulations in the isobaric-isothermal ensemble using hydrated dimethylphosphate anions (DMP(-)) and calcium cations. Results from the study demonstrate, formation of DMP-Ca(2+) complexes and the consequent removal of water, supporting the hypothesis. Our study further demonstrates that as a result of Ca(2+)-DMP self-assembly, the distance between anionic oxygens between the two DMP molecules is reduced to 2.92A, which is in close agreement with the 2.8A SNARE-induced apposition established between opposing bilayers, reported earlier from X-ray diffraction measurements.
Figures








Comment in
-
Membrane fusion--new light on the atomic organization of ions and molecules at the inter-bilayer leading to dehydration.Cell Biol Int. 2008 Apr;32(4):359-60. doi: 10.1016/j.cellbi.2008.03.003. Epub 2008 Mar 20. Cell Biol Int. 2008. PMID: 18452808 No abstract available.
References
-
- Bockmann RA, Grubmuller H. Multistep binding of divalent cations to phospholipid bilayers: a molecular dynamics study. Angew. Chem. Int. Ed. 2004;43:1021–1024. - PubMed
-
- Foloppe N, MacKerell AD., Jr An improved empirical potential energy function for molecular simulations of phospholipids. J. Comput. Chem. 2000;21:86–104.
-
- Guan Y, Choy GS-C, Glaser R, Thomas G., Jr Vibrational Analysis of Nucleic Acids. 2. Ab Initio Calculation of the Molecular Force Field and Normal Modes of Dimethyl Phosphate. J. Phys. Chem. 1995;99:12054–12062.
-
- Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Molec. Graphics. 1996;14:33–38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

