Consequences of C4 differentiation for chloroplast membrane proteomes in maize mesophyll and bundle sheath cells
- PMID: 18453340
- PMCID: PMC2556027
- DOI: 10.1074/mcp.M800016-MCP200
Consequences of C4 differentiation for chloroplast membrane proteomes in maize mesophyll and bundle sheath cells
Abstract
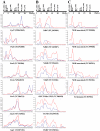
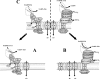
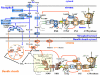
Chloroplasts of maize leaves differentiate into specific bundle sheath (BS) and mesophyll (M) types to accommodate C(4) photosynthesis. Chloroplasts contain thylakoid and envelope membranes that contain the photosynthetic machineries and transporters but also proteins involved in e.g. protein homeostasis. These chloroplast membranes must be specialized within each cell type to accommodate C(4) photosynthesis and regulate metabolic fluxes and activities. This quantitative study determined the differentiated state of BS and M chloroplast thylakoid and envelope membrane proteomes and their oligomeric states using innovative gel-based and mass spectrometry-based protein quantifications. This included native gels, iTRAQ, and label-free quantification using an LTQ-Orbitrap. Subunits of Photosystems I and II, the cytochrome b(6)f, and ATP synthase complexes showed average BS/M accumulation ratios of 1.6, 0.45, 1.0, and 1.33, respectively, whereas ratios for the light-harvesting complex I and II families were 1.72 and 0.68, respectively. A 1000-kDa BS-specific NAD(P)H dehydrogenase complex with associated proteins of unknown function containing more than 15 proteins was observed; we speculate that this novel complex possibly functions in inorganic carbon concentration when carboxylation rates by ribulose-bisphosphate carboxylase/oxygenase are lower than decarboxylation rates by malic enzyme. Differential accumulation of thylakoid proteases (Egy and DegP), state transition kinases (STN7,8), and Photosystem I and II assembly factors was observed, suggesting that cell-specific photosynthetic electron transport depends on post-translational regulatory mechanisms. BS/M ratios for inner envelope transporters phosphoenolpyruvate/P(i) translocator, Dit1, Dit2, and Mex1 were determined and reflect metabolic fluxes in carbon metabolism. A wide variety of hundreds of other proteins showed differential BS/M accumulation. Mass spectral information and functional annotations are available through the Plant Proteome Database. These data are integrated with previous data, resulting in a model for C(4) photosynthesis, thereby providing new rationales for metabolic engineering of C(4) pathways and targeted analysis of genetic networks that coordinate C(4) differentiation.
Figures



References
-
- Sheen, J. ( 1999) C-4 gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 187–217 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

