The rice StMADS11-like genes OsMADS22 and OsMADS47 cause floral reversions in Arabidopsis without complementing the svp and agl24 mutants
- PMID: 18453531
- PMCID: PMC2413287
- DOI: 10.1093/jxb/ern083
The rice StMADS11-like genes OsMADS22 and OsMADS47 cause floral reversions in Arabidopsis without complementing the svp and agl24 mutants
Abstract
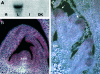
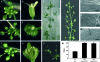
During floral induction and flower development plants undergo delicate phase changes which are under tight molecular control. MADS-box transcription factors have been shown to play pivotal roles during these transition phases. SHORT VEGETATIVE PHASE (SVP) and AGAMOUS LIKE 24 (AGL24) are important regulators both during the transition to flowering and during flower development. During vegetative growth they exert opposite roles on floral transition, acting as repressor and promoter of flowering, respectively. Later during flower development they act redundantly as negative regulators of AG expression. In rice, the orthologues of SVP and AGL24 are OsMADS22, OsMADS47, and OsMADS55 and these three genes are involved in the negative regulation of brassinosteroid responses. In order to understand whether these rice genes have maintained the ability to function as regulators of flowering time in Arabidopsis, complementation tests were performed by expressing OsMADS22 and OsMADS47 in the svp and agl24 mutants. The results show that the rice genes are not able to complement the flowering-time phenotype of the Arabidopsis mutants, indicating that they are biologically inactive in Arabidopsis. Nevertheless, they cause floral reversions, which mimic the SVP and AGL24 floral overexpressor phenotypes. Yeast two-hybrid analysis suggests that these floral phenotypes are probably the consequence of protein interactions between OsMADS22 and OsMADS47 and other MADS-box proteins which interfere with the formation of complexes required for normal flower development.
Figures



Similar articles
-
Functional conservation and diversification between rice OsMADS22/OsMADS55 and Arabidopsis SVP proteins.Plant Sci. 2012 Apr;185-186:97-104. doi: 10.1016/j.plantsci.2011.09.003. Epub 2011 Sep 12. Plant Sci. 2012. PMID: 22325870
-
AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis.Plant Cell. 2006 Jun;18(6):1373-82. doi: 10.1105/tpc.106.041798. Epub 2006 May 5. Plant Cell. 2006. PMID: 16679456 Free PMC article.
-
The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes.Plant J. 2009 Nov;60(4):626-37. doi: 10.1111/j.1365-313X.2009.03985.x. Epub 2009 Jul 25. Plant J. 2009. PMID: 19656343
-
Regulation and function of SOC1, a flowering pathway integrator.J Exp Bot. 2010 May;61(9):2247-54. doi: 10.1093/jxb/erq098. Epub 2010 Apr 22. J Exp Bot. 2010. PMID: 20413527 Review.
-
Make hay when the sun shines: the role of MADS-box genes in temperature-dependant seasonal flowering responses.Plant Sci. 2011 Mar;180(3):447-53. doi: 10.1016/j.plantsci.2010.12.001. Epub 2010 Dec 14. Plant Sci. 2011. PMID: 21421391 Review.
Cited by
-
Overexpression of PvSVP1, an SVP-like gene of bamboo, causes early flowering and abnormal floral organs in Arabidopsis and rice.Acta Biochim Biophys Sin (Shanghai). 2023 Jan 25;55(2):237-249. doi: 10.3724/abbs.2022199. Acta Biochim Biophys Sin (Shanghai). 2023. PMID: 36647724 Free PMC article.
-
The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals.Ann Bot. 2009 Jun;103(8):1165-72. doi: 10.1093/aob/mcp063. Epub 2009 Mar 21. Ann Bot. 2009. PMID: 19304997 Free PMC article. Review.
-
Evolution of major flowering pathway integrators in Orchidaceae.Plant Reprod. 2024 Jun;37(2):85-109. doi: 10.1007/s00497-023-00482-7. Epub 2023 Oct 12. Plant Reprod. 2024. PMID: 37823912 Free PMC article.
-
Identification and characterization of RcMADS1, an AGL24 ortholog from the holoparasitic plant Rafflesia cantleyi Solms-Laubach (Rafflesiaceae).PLoS One. 2013 Jun 28;8(6):e67243. doi: 10.1371/journal.pone.0067243. Print 2013. PLoS One. 2013. PMID: 23840638 Free PMC article.
-
Characterization and Expression Analysis of PtAGL24, a SHORT VEGETATIVE PHASE/AGAMOUS-LIKE 24 (SVP/AGL24)-Type MADS-Box Gene from Trifoliate Orange (Poncirus trifoliata L. Raf.).Front Plant Sci. 2016 Jun 10;7:823. doi: 10.3389/fpls.2016.00823. eCollection 2016. Front Plant Sci. 2016. PMID: 27375669 Free PMC article.
References
-
- Becker A, Theissen G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Molecular Phylogenetic Evolution. 2003;29:464–489. - PubMed
-
- Carmona MJ, Ortega N, Garcia-Maroto F. Isolation and molecular characterization of a new vegetative MADS-box gene from Solanum tuberosum L. Planta. 1998;207:181–188. - PubMed
-
- Ciannamea S, Kaufmann K, Frau M, Tonaco IA, Petersen K, Nielsen KK, Angenent GC, Immink RG. Protein interactions of MADS box transcription factors involved in flowering in Lolium perenne. Journal of Experimental Botany. 2006;57:3419–3431. - PubMed

