Structure of a rabbit muscle fructose-1,6-bisphosphate aldolase A dimer variant
- PMID: 18453690
- PMCID: PMC2631105
- DOI: 10.1107/S0907444908004976
Structure of a rabbit muscle fructose-1,6-bisphosphate aldolase A dimer variant
Abstract
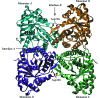
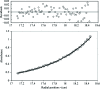
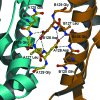
Fructose-1,6-bisphosphate aldolase (aldolase) is an essential enzyme in glycolysis and gluconeogenesis. In addition to this primary function, aldolase is also known to bind to a variety of other proteins, a property that may allow it to perform 'moonlighting' roles in the cell. Although monomeric and dimeric aldolases possess full catalytic activity, the enzyme occurs as an unusually stable tetramer, suggesting a possible link between the oligomeric state and these noncatalytic cellular roles. Here, the first high-resolution X-ray crystal structure of rabbit muscle D128V aldolase, a dimeric form of aldolase mimicking the clinically important D128G mutation in humans associated with hemolytic anemia, is presented. The structure of the dimer was determined to 1.7 angstroms resolution with the product DHAP bound in the active site. The turnover of substrate to produce the product ligand demonstrates the retention of catalytic activity by the dimeric aldolase. The D128V mutation causes aldolase to lose intermolecular contacts with the neighboring subunit at one of the two interfaces of the tetramer. The tertiary structure of the dimer does not significantly differ from the structure of half of the tetramer. Analytical ultracentrifugation confirms the occurrence of the enzyme as a dimer in solution. The highly stable structure of aldolase with an independent active site is consistent with a model in which aldolase has evolved as a multimeric scaffold to perform other noncatalytic functions.
Figures

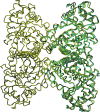
Similar articles
-
Subunit interface mutants of rabbit muscle aldolase form active dimers.Protein Sci. 1994 Sep;3(9):1383-91. doi: 10.1002/pro.5560030904. Protein Sci. 1994. PMID: 7833800 Free PMC article.
-
Snapshots of catalysis: the structure of fructose-1,6-(bis)phosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate.Biochemistry. 2001 Nov 20;40(46):13868-75. doi: 10.1021/bi0114877. Biochemistry. 2001. PMID: 11705376
-
Exploring substrate binding and discrimination in fructose1, 6-bisphosphate and tagatose 1,6-bisphosphate aldolases.Eur J Biochem. 2000 Mar;267(6):1858-68. doi: 10.1046/j.1432-1327.2000.01191.x. Eur J Biochem. 2000. PMID: 10712619
-
Structure, function and evolution of the Archaeal class I fructose-1,6-bisphosphate aldolase.Biochem Soc Trans. 2004 Apr;32(Pt 2):259-63. doi: 10.1042/bst0320259. Biochem Soc Trans. 2004. PMID: 15046584 Review.
-
Multifunctional Fructose 1,6-Bisphosphate Aldolase as a Therapeutic Target.Front Mol Biosci. 2021 Aug 11;8:719678. doi: 10.3389/fmolb.2021.719678. eCollection 2021. Front Mol Biosci. 2021. PMID: 34458323 Free PMC article. Review.
Cited by
-
"Hot standards" for the thermoacidophilic archaeon Sulfolobus solfataricus.Extremophiles. 2010 Jan;14(1):119-42. doi: 10.1007/s00792-009-0280-0. Epub 2009 Oct 4. Extremophiles. 2010. PMID: 19802714 Free PMC article.
-
Proteinaceous secretion of bioadhesive produced during crawling and settlement of Crassostrea gigas larvae.Sci Rep. 2018 Oct 17;8(1):15298. doi: 10.1038/s41598-018-33720-4. Sci Rep. 2018. PMID: 30333557 Free PMC article.
-
Molecular Characterization, Gene Evolution, and Expression Analysis of the Fructose-1, 6-bisphosphate Aldolase (FBA) Gene Family in Wheat (Triticum aestivum L.).Front Plant Sci. 2017 Jun 14;8:1030. doi: 10.3389/fpls.2017.01030. eCollection 2017. Front Plant Sci. 2017. PMID: 28659962 Free PMC article.
-
Aldolase A accelerates hepatocarcinogenesis by refactoring c-Jun transcription.J Pharm Anal. 2025 Jul;15(7):101169. doi: 10.1016/j.jpha.2024.101169. Epub 2024 Dec 16. J Pharm Anal. 2025. PMID: 40708574 Free PMC article.
-
Automated cryo-EM structure refinement using correlation-driven molecular dynamics.Elife. 2019 Mar 4;8:e43542. doi: 10.7554/eLife.43542. Elife. 2019. PMID: 30829573 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

