Identification of small molecule inhibitors of Pseudomonas aeruginosa exoenzyme S using a yeast phenotypic screen
- PMID: 18454192
- PMCID: PMC2265467
- DOI: 10.1371/journal.pgen.1000005
Identification of small molecule inhibitors of Pseudomonas aeruginosa exoenzyme S using a yeast phenotypic screen
Erratum in
- PLoS Genet. 2008 Apr;4(4). doi: 10.1371/annotation/76d35829-07a2-479f-bbc1-cce6755b6d8c. Merrill, Rod A [corrected to Merrill, A Rod] doi: 10.1371/annotation/76d35829-07a2-479f-bbc1-cce6755b6d8c
Abstract
Pseudomonas aeruginosa is an opportunistic human pathogen that is a key factor in the mortality of cystic fibrosis patients, and infection represents an increased threat for human health worldwide. Because resistance of Pseudomonas aeruginosa to antibiotics is increasing, new inhibitors of pharmacologically validated targets of this bacterium are needed. Here we demonstrate that a cell-based yeast phenotypic assay, combined with a large-scale inhibitor screen, identified small molecule inhibitors that can suppress the toxicity caused by heterologous expression of selected Pseudomonas aeruginosa ORFs. We identified the first small molecule inhibitor of Exoenzyme S (ExoS), a toxin involved in Type III secretion. We show that this inhibitor, exosin, modulates ExoS ADP-ribosyltransferase activity in vitro, suggesting the inhibition is direct. Moreover, exosin and two of its analogues display a significant protective effect against Pseudomonas infection in vivo. Furthermore, because the assay was performed in yeast, we were able to demonstrate that several yeast homologues of the known human ExoS targets are likely ADP-ribosylated by the toxin. For example, using an in vitro enzymatic assay, we demonstrate that yeast Ras2p is directly modified by ExoS. Lastly, by surveying a collection of yeast deletion mutants, we identified Bmh1p, a yeast homologue of the human FAS, as an ExoS cofactor, revealing that portions of the bacterial toxin mode of action are conserved from yeast to human. Taken together, our integrated cell-based, chemical-genetic approach demonstrates that such screens can augment traditional drug screening approaches and facilitate the discovery of new compounds against a broad range of human pathogens.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

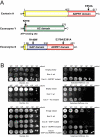

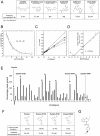

References
-
- Furuya EY, Lowy FD. Antimicrobial-resistant bacteria in the community setting. Nat Rev Microbiol. 2006;4:36–45. - PubMed
-
- Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. 2004;10:S122–129. - PubMed
-
- Fernandes P. Antibacterial discovery and development–the failure of success? Nat Biotechnol. 2006;24:1497–1503. - PubMed
-
- Christoffersen RE. Antibiotics–an investment worth making? Nat Biotechnol. 2006;24:1512–1514. - PubMed
-
- Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

