Two-state allosteric modeling suggests protein equilibrium as an integral component for cyclic AMP (cAMP) specificity in the cAMP receptor protein of Escherichia coli
- PMID: 18456811
- PMCID: PMC2446778
- DOI: 10.1128/JB.00074-08
Two-state allosteric modeling suggests protein equilibrium as an integral component for cyclic AMP (cAMP) specificity in the cAMP receptor protein of Escherichia coli
Abstract
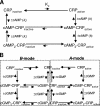
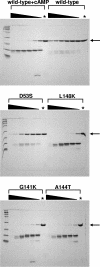
Activation of the cAMP receptor protein (CRP) from Escherichia coli is highly specific to its allosteric ligand, cAMP. Ligands such as adenosine and cGMP, which are structurally similar to cAMP, fail to activate wild-type CRP. However, several cAMP-independent CRP variants (termed CRP*) exist that can be further activated by both adenosine and cGMP, as well as by cAMP. This has remained a puzzle because the substitutions in many of these CRP* variants lie far from the cAMP-binding pocket (>10 A) and therefore should not directly affect that pocket. Here we show a surprising similarity in the altered ligand specificity of four CRP* variants with a single substitution in D53S, G141K, A144T, or L148K, and we propose a common basis for this phenomenon. The increased active protein population caused by an equilibrium shift in these variants is hypothesized to preferentially stabilize ligand binding. This explanation is completely consistent with the cAMP specificity in the activation of wild-type CRP. The model also predicts that wild-type CRP should be activated even by the lower-affinity ligand, adenosine, which we experimentally confirmed. The study demonstrates that protein equilibrium is an integral factor for ligand specificity in an allosteric protein, in addition to the direct effects of ligand pocket residues.
Figures


References
-
- Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293199-213. - PubMed
-
- Changeux, J.-P., and S. J. Edelstein. 2005. Allosteric mechanisms of signal transduction. Science 3081424-1428. - PubMed
-
- Cheng, X., and J. C. Lee. 1994. Absolute requirement of cyclic nucleotide in the activation of the G141Q mutant cAMP receptor protein from Escherichia coli. J. Biol. Chem. 26930781-30784. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

