Acyl chain specificity of the acyltransferases LpxA and LpxD and substrate availability contribute to lipid A fatty acid heterogeneity in Porphyromonas gingivalis
- PMID: 18456814
- PMCID: PMC2446808
- DOI: 10.1128/JB.00234-08
Acyl chain specificity of the acyltransferases LpxA and LpxD and substrate availability contribute to lipid A fatty acid heterogeneity in Porphyromonas gingivalis
Abstract
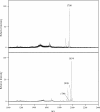
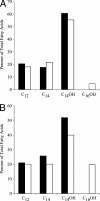
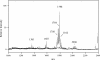
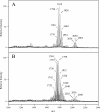
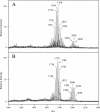
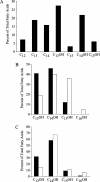
Porphyromonas gingivalis lipid A is heterogeneous with regard to the number, type, and placement of fatty acids. Analysis of lipid A by matrix-assisted laser desorption ionization-time of flight mass spectrometry reveals clusters of peaks differing by 14 mass units indicative of an altered distribution of the fatty acids generating different lipid A structures. To examine whether the transfer of hydroxy fatty acids with different chain lengths could account for the clustering of lipid A structures, P. gingivalis lpxA (lpxA(Pg)) and lpxD(Pg) were cloned and expressed in Escherichia coli strains in which the homologous gene was mutated. Lipid A from strains expressing either of the P. gingivalis transferases was found to contain 16-carbon hydroxy fatty acids in addition to the normal E. coli 14-carbon hydroxy fatty acids, demonstrating that these acyltransferases display a relaxed acyl chain length specificity. Both LpxA and LpxD, from either E. coli or P. gingivalis, were also able to incorporate odd-chain fatty acids into lipid A when grown in the presence of 1% propionic acid. This indicates that E. coli lipid A acyltransferases do not have an absolute specificity for 14-carbon hydroxy fatty acids but can transfer fatty acids differing by one carbon unit if the fatty acid substrates are available. We conclude that the relaxed specificity of the P. gingivalis lipid A acyltransferases and the substrate availability account for the lipid A structural clusters that differ by 14 mass units observed in P. gingivalis lipopolysaccharide preparations.
Figures


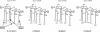
Similar articles
-
Expression of a Porphyromonas gingivalis lipid A palmitylacyltransferase in Escherichia coli yields a chimeric lipid A with altered ability to stimulate interleukin-8 secretion.Cell Microbiol. 2006 Jan;8(1):120-9. doi: 10.1111/j.1462-5822.2005.00605.x. Cell Microbiol. 2006. PMID: 16367871
-
Relaxed acyl chain specificity of Bordetella UDP-N-acetylglucosamine acyltransferases.J Biol Chem. 2002 May 24;277(21):18281-90. doi: 10.1074/jbc.M201057200. Epub 2002 Mar 11. J Biol Chem. 2002. PMID: 11889134
-
Structural basis for the recognition of peptide RJPXD33 by acyltransferases in lipid A biosynthesis.J Biol Chem. 2014 May 30;289(22):15527-35. doi: 10.1074/jbc.M114.564278. Epub 2014 Apr 16. J Biol Chem. 2014. PMID: 24742680 Free PMC article.
-
Chemical structure and immunobiological activity of Porphyromonas gingivalis lipid A.Front Biosci. 2007 May 1;12:3795-812. doi: 10.2741/2353. Front Biosci. 2007. PMID: 17485340 Review.
-
Acyltransferases in bacteria.Microbiol Mol Biol Rev. 2013 Jun;77(2):277-321. doi: 10.1128/MMBR.00010-13. Microbiol Mol Biol Rev. 2013. PMID: 23699259 Free PMC article. Review.
Cited by
-
Prediction of Selected Biosynthetic Pathways for the Lipopolysaccharide Components in Porphyromonas gingivalis.Pathogens. 2021 Mar 20;10(3):374. doi: 10.3390/pathogens10030374. Pathogens. 2021. PMID: 33804654 Free PMC article.
-
Lipid A Structural Divergence in Rickettsia Pathogens.mSphere. 2021 May 5;6(3):e00184-21. doi: 10.1128/mSphere.00184-21. mSphere. 2021. PMID: 33952661 Free PMC article.
-
A pathway-directed positive growth restoration assay to facilitate the discovery of lipid A and fatty acid biosynthesis inhibitors in Acinetobacter baumannii.PLoS One. 2018 Mar 5;13(3):e0193851. doi: 10.1371/journal.pone.0193851. eCollection 2018. PLoS One. 2018. PMID: 29505586 Free PMC article.
-
Kdo2 -lipid A: structural diversity and impact on immunopharmacology.Biol Rev Camb Philos Soc. 2015 May;90(2):408-27. doi: 10.1111/brv.12114. Epub 2014 May 16. Biol Rev Camb Philos Soc. 2015. PMID: 24838025 Free PMC article. Review.
-
Rickettsia Lipid A Biosynthesis Utilizes the Late Acyltransferase LpxJ for Secondary Fatty Acid Addition.J Bacteriol. 2018 Sep 10;200(19):e00334-18. doi: 10.1128/JB.00334-18. Print 2018 Oct 1. J Bacteriol. 2018. PMID: 30012728 Free PMC article.
References
-
- Amano, A., I. Nakagawa, N. Okahashi, and N. Hamada. 2004. Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J. Periodontal Res. 39136-142. - PubMed
-
- Bainbridge, B., and R. Darveau. 1999. Role of LPS from oral bacteria in health and disease, p. 899-914. In H. Brade, S. M. Opal, S. Vogel, and D. Morrison (ed.), Endotoxin in health and disease. Marcel Dekker, New York, NY.
-
- Bainbridge, B. W., S. R. Coats, T. T. Pham, R. A. Reife, and R. P. Darveau. 2006. Expression of a Porphyromonas gingivalis lipid A palmitylacyltransferase in Escherichia coli yields a chimeric lipid A with altered ability to stimulate interleukin-8 secretion. Cell Microbiol. 8120-129. - PubMed
-
- Bainbridge, B. W., and R. P. Darveau. 2001. Porphyromonas gingivalis lipopolysaccharide: an unusual pattern recognition receptor ligand for the innate host defense system. Acta Odontol. Scand. 59131-138. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

