Monte Carlo study of single molecule diffusion can elucidate the mechanism of B cell synapse formation
- PMID: 18456833
- PMCID: PMC2479588
- DOI: 10.1529/biophysj.107.122564
Monte Carlo study of single molecule diffusion can elucidate the mechanism of B cell synapse formation
Abstract
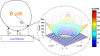
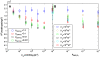
B cell receptors have been shown to cluster at the intercellular junction between a B cell and an antigen-presenting cell in the form of a segregated pattern of B cell receptor/antigen complexes known as an immunological synapse. We use random walk-based theoretical arguments and Monte Carlo simulations to study the effect of diffusion of surface-bound molecules on B cell synapse formation. Our results show that B cell synapse formation is optimal for a limited range of receptor-ligand complex diffusion coefficient values, typically one-to-two orders of magnitude lower than the diffusion coefficient of free receptors. Such lower mobility of receptor-ligand complexes can significantly affect the diffusion of a tagged receptor or ligand in an affinity dependent manner, as the binding/unbinding of such receptor or ligand molecules crucially depends on affinity. Our work shows how single molecule tracking experiments can be used to estimate the order of magnitude of the diffusion coefficient of receptor-ligand complexes, which is difficult to measure directly in experiments due to the finite lifetime of receptor-ligand bonds. We also show how such antigen movement data at the single molecule level can provide insight into the B cell synapse formation mechanism. Thus, our results can guide further single molecule tracking experiments to elucidate the synapse formation mechanism in B cells, and potentially in other immune cells.
Figures




References
-
- Lauffenburger, D. A., and J. J. Linderman. 1993. Models for binding, trafficking and signaling. Oxford University Press, Oxford.
-
- Monks, C. R., B. A. Freiberg, H. Kupfer, N. Sciaky, and A. Kupfer. 1998. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 395:82–86. - PubMed
-
- Grakoui, A., S. K. Bromley, C. Sumen, M. M. Davis, A. S. Shaw, P. M. Allen, and M. L. Dustin. 1999. The immunological synapse: a molecular machine controlling T cell activation. Science. 285:221–227. - PubMed
-
- Krummel, M. F., M. D. Sjaastad, C. Wulfing, and M. M. Davis. 2000. Differential clustering of CD4 and CD3ζ during T cell recognition. Science. 289:1349–1352. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

