Influence of genotype and nutrition on survival and metabolism of starving yeast
- PMID: 18456835
- PMCID: PMC2383937
- DOI: 10.1073/pnas.0802601105
Influence of genotype and nutrition on survival and metabolism of starving yeast
Abstract
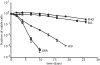
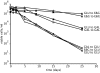
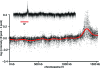
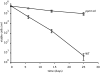
Starvation of yeast cultures limited by auxotrophic requirements results in glucose wasting and failure to achieve prompt cell-cycle arrest when compared with starvation for basic natural nutrients like phosphate or sulfate. We measured the survival of yeast auxotrophs upon starvation for different nutrients and found substantial differences: When deprived of leucine or uracil, viability declined exponentially with a half-life of <2 days, whereas when the same strains were deprived of phosphate or sulfate, the half-life was approximately 10 days. The survival rates of nongrowing auxotrophs deprived of uracil or leucine depended on the carbon source available during starvation, but were independent of the carbon source during prior growth. We performed an enrichment procedure for mutants that suppress lethality during auxotrophy starvation. We repeatedly found loss-of-function mutations in a number of functionally related genes. Mutations in PPM1, which methylates protein phosphatase 2A, and target of rapamycin (TOR1) were characterized further. Deletion of PPM1 almost completely suppressed the rapid lethality and substantially suppressed glucose wasting during starvation for leucine or uracil. Suppression by a deletion of TOR1 was less complete. We suggest that, similar to the Warburg effect observed in tumor cells, starving yeast auxotrophs wastes glucose as a consequence of the failure of conserved growth control pathways. Furthermore, we suggest that our results on condition-dependent chronological lifespan have important implications for the interpretation and design of studies on chronological aging.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Boer VM, de Winde JH, Pronk JT, Piper MD. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J Biol Chem. 2003;278:3265–3274. - PubMed
-
- Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. - PubMed
-
- Schneper L, Duvel K, Broach JR. Sense and sensibility: Nutritional response and signal integration in yeast. Curr Opin Microbiol. 2004;7:624–630. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

