Ab initio RNA folding by discrete molecular dynamics: from structure prediction to folding mechanisms
- PMID: 18456842
- PMCID: PMC2390798
- DOI: 10.1261/rna.894608
Ab initio RNA folding by discrete molecular dynamics: from structure prediction to folding mechanisms
Abstract
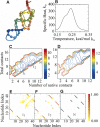
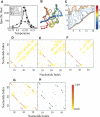
RNA molecules with novel functions have revived interest in the accurate prediction of RNA three-dimensional (3D) structure and folding dynamics. However, existing methods are inefficient in automated 3D structure prediction. Here, we report a robust computational approach for rapid folding of RNA molecules. We develop a simplified RNA model for discrete molecular dynamics (DMD) simulations, incorporating base-pairing and base-stacking interactions. We demonstrate correct folding of 150 structurally diverse RNA sequences. The majority of DMD-predicted 3D structures have <4 A deviations from experimental structures. The secondary structures corresponding to the predicted 3D structures consist of 94% native base-pair interactions. Folding thermodynamics and kinetics of tRNA(Phe), pseudoknots, and mRNA fragments in DMD simulations are in agreement with previous experimental findings. Folding of RNA molecules features transient, non-native conformations, suggesting non-hierarchical RNA folding. Our method allows rapid conformational sampling of RNA folding, with computational time increasing linearly with RNA length. We envision this approach as a promising tool for RNA structural and functional analyses.
Figures



Similar articles
-
iFoldRNA: three-dimensional RNA structure prediction and folding.Bioinformatics. 2008 Sep 1;24(17):1951-2. doi: 10.1093/bioinformatics/btn328. Epub 2008 Jun 25. Bioinformatics. 2008. PMID: 18579566 Free PMC article.
-
RNA folding on the 3D triangular lattice.BMC Bioinformatics. 2009 Nov 5;10:369. doi: 10.1186/1471-2105-10-369. BMC Bioinformatics. 2009. PMID: 19891777 Free PMC article.
-
Ab initio folding of proteins with all-atom discrete molecular dynamics.Structure. 2008 Jul;16(7):1010-8. doi: 10.1016/j.str.2008.03.013. Structure. 2008. PMID: 18611374 Free PMC article.
-
Prediction of RNA secondary structure by free energy minimization.Curr Opin Struct Biol. 2006 Jun;16(3):270-8. doi: 10.1016/j.sbi.2006.05.010. Epub 2006 May 19. Curr Opin Struct Biol. 2006. PMID: 16713706 Review.
-
Ab initio RNA folding.J Phys Condens Matter. 2015 Jun 17;27(23):233102. doi: 10.1088/0953-8984/27/23/233102. Epub 2015 May 20. J Phys Condens Matter. 2015. PMID: 25993396 Review.
Cited by
-
Modeling Structure, Stability, and Flexibility of Double-Stranded RNAs in Salt Solutions.Biophys J. 2018 Oct 16;115(8):1403-1416. doi: 10.1016/j.bpj.2018.08.030. Epub 2018 Aug 30. Biophys J. 2018. PMID: 30236782 Free PMC article.
-
RNA Bricks--a database of RNA 3D motifs and their interactions.Nucleic Acids Res. 2014 Jan;42(Database issue):D123-31. doi: 10.1093/nar/gkt1084. Epub 2013 Nov 12. Nucleic Acids Res. 2014. PMID: 24220091 Free PMC article.
-
A boost for the emerging field of RNA nanotechnology.ACS Nano. 2011 May 24;5(5):3405-18. doi: 10.1021/nn200989r. ACS Nano. 2011. PMID: 21604810 Free PMC article.
-
SimRNA: a coarse-grained method for RNA folding simulations and 3D structure prediction.Nucleic Acids Res. 2016 Apr 20;44(7):e63. doi: 10.1093/nar/gkv1479. Epub 2015 Dec 19. Nucleic Acids Res. 2016. PMID: 26687716 Free PMC article.
-
Exploring RNA conformational space under sparse distance restraints.Sci Rep. 2017 Mar 10;7:44074. doi: 10.1038/srep44074. Sci Rep. 2017. PMID: 28281575 Free PMC article.
References
-
- Andersen H.C. Molecular-dynamics simulations at constant pressure and-or temperature. J. Chem. Phys. 1980;72:2384–2393.
-
- Baumgartner A. Applications of the Monte-Carlo simulations in statistical physics. Springer; New York: 1987.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
