Extracellular plasma RNA from colon cancer patients is confined in a vesicle-like structure and is mRNA-enriched
- PMID: 18456845
- PMCID: PMC2441977
- DOI: 10.1261/rna.755908
Extracellular plasma RNA from colon cancer patients is confined in a vesicle-like structure and is mRNA-enriched
Abstract
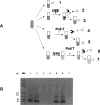
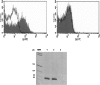
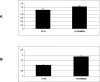
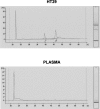
Little is yet known about the origin and protective mechanism of free nucleic acids in plasma. We investigated the possibility of these free nucleic acids being particle associated. Plasma samples from colon cancer patients and cell culture media were subjected to various antibody incubations, ultracentrifugation, and RNA extraction protocols for total RNA, epithelial RNA, and mRNA. Flow cytometry using a Ber-EP4 antibody and confocal laser microscopy after staining with propidium iodide were also performed. mRNA levels of the LISCH7 and SDHA genes were determined in cells and in culture media. Ber-EP4 antibody and polystyrene beads coated with oligo dT sequences were employed. We observed that, after incubation, total RNA and mRNA were always detected after membrane digestion, and that epithelial RNA was detected before this procedure. In ultracentrifugation, mRNA was caught in the supernatant only if a former lysis mediated or in the pellet if there was no previous digestion. Flow cytometry determinations showed that antibody-coated microbeads keep acellular structures bearing epithelial antigens apart. Confocal laser microscopy made 1- to 2-microm-diameter particles perceptible in the vicinity of magnetic polystyrene beads. Relevant differences were observed between mRNA of cells and culture media, as there was a considerable difference in LISCH7 mRNA levels between HT29 and IMR90 cell co-cultures and their culture media. Our results support the view that extracellular RNA found in plasma from cancer patients circulates in association with or is protected in a multiparticle complex, and that an active release mechanism by tumor cells may be a possible origin.
Figures


References
-
- Anker P., Stroun M., Maurice P.A. Spontaneous release of DNA by human blood lymphoctyes as shown in an in vitro system. Cancer Res. 1975;35:2375–2382. - PubMed
-
- Anker P., Lefort F., Vasioukhin V., Lyautey J., Lederrey C., Chen X.Q., Stroun M., Mulcahy H.E., Farthing M.J. K-ras mutations are found in DNA extracted from the plasma of patients with colorectal cancer. Gastroenterology. 1997;112:1114–1120. - PubMed
-
- Anker P., Mulcahy H., Chen X.Q., Stroun M. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev. 1999;18:65–73. - PubMed
-
- Bhowmick N.A., Chytil A., Plieth D., Gorska A.E., Dumont N., Shappell S., Washington M.K., Neilson E.G., Moses H.L. TGF-β signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004a;303:848–851. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
