Multilocus sequence typing of pathogenic Candida species
- PMID: 18456859
- PMCID: PMC2446668
- DOI: 10.1128/EC.00062-08
Multilocus sequence typing of pathogenic Candida species
Figures
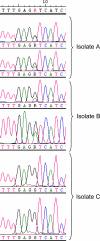
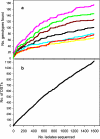
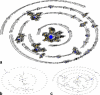
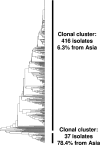
Similar articles
-
Use of multilocus sequence typing for the investigation of colonisation by Candida albicans in intensive care unit patients.J Hosp Infect. 2008 May;69(1):24-32. doi: 10.1016/j.jhin.2008.02.006. Epub 2008 Apr 8. J Hosp Infect. 2008. PMID: 18396349
-
Candida albicans or Candida dubliniensis?Mycoses. 2011 Jan;54(1):1-16. doi: 10.1111/j.1439-0507.2009.01759.x. Mycoses. 2011. PMID: 19682314 Review.
-
Identification of CARE-2-negative Candida albicans isolates as Candida dubliniensis.Mycoses. 1999 Apr;42(1-2):29-32. doi: 10.1046/j.1439-0507.1999.00259.x. Mycoses. 1999. PMID: 10394844
-
First molecular method for discriminating between Candida africana, Candida albicans, and Candida dubliniensis by using hwp1 gene.Diagn Microbiol Infect Dis. 2008 Oct;62(2):230-3. doi: 10.1016/j.diagmicrobio.2008.05.014. Epub 2008 Jul 21. Diagn Microbiol Infect Dis. 2008. PMID: 18640803
-
Molecular phylogenetics and epidemiology of Candida albicans.Future Microbiol. 2010 Jan;5(1):67-79. doi: 10.2217/fmb.09.113. Future Microbiol. 2010. PMID: 20020830 Review.
Cited by
-
Multilocus sequence typing of Candida albicans oral isolates reveals high genetic relatedness of mother-child dyads in early life.PLoS One. 2024 Jan 17;19(1):e0290938. doi: 10.1371/journal.pone.0290938. eCollection 2024. PLoS One. 2024. PMID: 38232064 Free PMC article.
-
Trailing or paradoxical growth of Candida albicans when exposed to caspofungin is not associated with microsatellite genotypes.Antimicrob Agents Chemother. 2010 Mar;54(3):1365-8. doi: 10.1128/AAC.00530-09. Epub 2010 Jan 11. Antimicrob Agents Chemother. 2010. PMID: 20065050 Free PMC article.
-
Candidemia surveillance in Brazil: evidence for a geographical boundary defining an area exhibiting an abatement of infections by Candida albicans group 2 strains.J Clin Microbiol. 2010 Sep;48(9):3062-7. doi: 10.1128/JCM.00262-10. Epub 2010 Jun 30. J Clin Microbiol. 2010. PMID: 20592158 Free PMC article.
-
Molecular Demonstration of a Pneumocystis Outbreak in Stem Cell Transplant Patients: Evidence for Transmission in the Daycare Center.Front Microbiol. 2017 Apr 24;8:700. doi: 10.3389/fmicb.2017.00700. eCollection 2017. Front Microbiol. 2017. PMID: 28484441 Free PMC article.
-
Property differences among the four major Candida albicans strain clades.Eukaryot Cell. 2009 Mar;8(3):373-87. doi: 10.1128/EC.00387-08. Epub 2009 Jan 16. Eukaryot Cell. 2009. PMID: 19151328 Free PMC article.
References
-
- Al Mosaid, A., D. J. Sullivan, I. Polacheck, F. A. Shaheen, O. Soliman, S. Al Hedaithy, S. Al Thawad, M. Kabadaya, and D. C. Coleman. 2005. Novel 5-flucytosine-resistant clade of Candida dubliniensis from Saudi Arabia and Egypt identified by Cd25 fingerprinting. J. Clin. Microbiol. 434026-4036. - PMC - PubMed
-
- Ball, L. M., M. A. Bes, B. Theelen, T. Boekhout, R. M. Egeler, and E. J. Kuijper. 2004. Significance of amplified fragment length polymorphism in identification and epidemiological examination of Candida species colonization in children undergoing allogeneic stem cell transplantation. J. Clin. Microbiol. 421673-1679. - PMC - PubMed
-
- Blignaut, E., J. Molepoa, C. Pujol, D. R. Soll, and M. A. Pfaller. 2005. Clade-related amphotericin B resistance among South African Candida albicans isolates. Diagn. Microbiol. Infect. Dis. 5329-31. - PubMed
-
- Bougnoux, M.-E., D. Diogo, N. Francois, B. Sendid, S. Veirmeire, J. F. Colombel, C. Bouchier, H. Van Kruiningen, C. d'Enfert, and D. Poulain. 2006. Multilocus sequence typing reveals intrafamilial transmission and microevolutions of Candida albicans isolates from the human digestive tract. J. Clin. Microbiol. 441810-1820. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

