DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts
- PMID: 18456864
- PMCID: PMC2493436
- DOI: 10.1101/gr.075713.107
DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts
Abstract
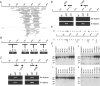
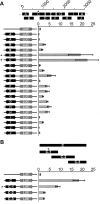
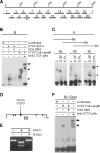
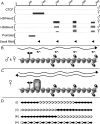
Macrosatellite DNA is composed of large repeat units, arranged in tandem over hundreds of kilobases. The macrosatellite repeat DXZ4, localized at Xq23-24, consists of 50-100 copies of a CpG-rich 3-kb monomer. Here I report that on the active X chromosome (Xa), DXZ4 is organized into constitutive heterochromatin characterized by a highly organized pattern of H3K9me3. DXZ4 is expressed from both strands and generates an antisense transcript that is processed into small RNAs that directly correlate with H3K9me3 nucleosomes. In contrast, on the inactive X chromosome (Xi) a proportion of DXZ4 is packaged into euchromatin characterized by H3K4me2 and H3K9Ac. The Xi copy of DXZ4 is bound by the chromatin insulator, CTCF, within a sequence that unidirectionally blocks enhancer-promoter communication. Immediately adjacent to the CTCF-binding site is a bidirectional promoter that, like the sequence flanking the CTCF-binding region, is completely devoid of CpG methylation on the Xi. As on the Xa, both strands are expressed, but longer antisense transcripts can be detected in addition to the processed small RNAs. The euchromatic organization of DXZ4 on the otherwise heterochromatic Xi, its binding of CTCF, and its function as a unidirectional insulator suggest that this macrosatellite has acquired a novel function unique to the process of X chromosome inactivation.
Figures




References
-
- Bell A.C., Felsenfeld G., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. - PubMed
-
- Bell A.C., West A.G., Felsenfeld G., West A.G., Felsenfeld G., Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. - PubMed
-
- Boggs B.A., Cheung P., Heard E., Spector D.L., Chinault A.C., Allis C.D., Cheung P., Heard E., Spector D.L., Chinault A.C., Allis C.D., Heard E., Spector D.L., Chinault A.C., Allis C.D., Spector D.L., Chinault A.C., Allis C.D., Chinault A.C., Allis C.D., Allis C.D. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 2002;30:73–76. - PubMed
-
- Boumil R.M., Ogawa Y., Sun B.K., Huynh K.D., Lee J.T., Ogawa Y., Sun B.K., Huynh K.D., Lee J.T., Sun B.K., Huynh K.D., Lee J.T., Huynh K.D., Lee J.T., Lee J.T. Differential methylation of Xite and CTCF sites in Tsix mirrors the pattern of X-inactivation choice in mice. Mol. Cell. Biol. 2006;26:2109–2117. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
