Understanding the development of human bladder cancer by using a whole-organ genomic mapping strategy
- PMID: 18458673
- PMCID: PMC2849658
- DOI: 10.1038/labinvest.2008.27
Understanding the development of human bladder cancer by using a whole-organ genomic mapping strategy
Abstract
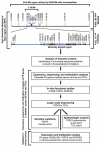
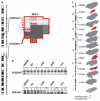
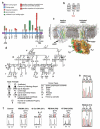
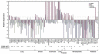
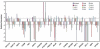
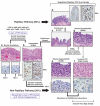
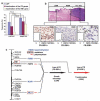
The search for the genomic sequences involved in human cancers can be greatly facilitated by maps of genomic imbalances identifying the involved chromosomal regions, particularly those that participate in the development of occult preneoplastic conditions that progress to clinically aggressive invasive cancer. The integration of such regions with human genome sequence variation may provide valuable clues about their overall structure and gene content. By extension, such knowledge may help us understand the underlying genetic components involved in the initiation and progression of these cancers. We describe the development of a genome-wide map of human bladder cancer that tracks its progression from in situ precursor conditions to invasive disease. Testing for allelic losses using a genome-wide panel of 787 microsatellite markers was performed on multiple DNA samples, extracted from the entire mucosal surface of the bladder and corresponding to normal urothelium, in situ preneoplastic lesions, and invasive carcinoma. Using this approach, we matched the clonal allelic losses in distinct chromosomal regions to specific phases of bladder neoplasia and produced a detailed genetic map of bladder cancer development. These analyses revealed three major waves of genetic changes associated with growth advantages of successive clones and reflecting a stepwise conversion of normal urothelial cells into cancer cells. The genetic changes map to six regions at 3q22-q24, 5q22-q31, 9q21-q22, 10q26, 13q14, and 17p13, which may represent critical hits driving the development of bladder cancer. Finally, we performed high-resolution mapping using single nucleotide polymorphism markers within one region on chromosome 13q14, containing the model tumor suppressor gene RB1, and defined a minimal deleted region associated with clonal expansion of in situ neoplasia. These analyses provided new insights on the involvement of several non-coding sequences mapping to the region and identified novel target genes, termed forerunner (FR) genes, involved in early phases of cancer development.
Figures









References
-
- Ayala AG, Ro JY, Amin M, editors. Pathology of Incipient Neoplesia. Oxford University Press; New York: 2001.
-
- Jemal A, Siegel R, Ward E, et al. Cancer statistics. CA Cancer J Clin. 2007;57:43–66. - PubMed
-
- Lynch CF, Cohen MB. Urinary system. Cancer. 1995;75(1 Suppl):316–329. - PubMed
-
- Dinney CP, McConkey DJ, Millikan RE, et al. Focus on bladder cancer. Cancer Cell. 2004;6:111–116. - PubMed
-
- Spiess PE, Czerniak B. Dual-track pathway of bladder carcinogenesis: practical implications. Arch Pathol Lab Med. 2006;130:844–852. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

