Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer
- PMID: 18462018
- PMCID: PMC2365982
- DOI: 10.1371/journal.pbio.0060108
Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer
Abstract
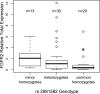
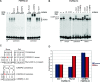
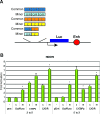
The recent whole-genome scan for breast cancer has revealed the FGFR2 (fibroblast growth factor receptor 2) gene as a locus associated with a small, but highly significant, increase in the risk of developing breast cancer. Using fine-scale genetic mapping of the region, it has been possible to narrow the causative locus to a haplotype of eight strongly linked single nucleotide polymorphisms (SNPs) spanning a region of 7.5 kilobases (kb) in the second intron of the FGFR2 gene. Here we describe a functional analysis to define the causative SNP, and we propose a model for a disease mechanism. Using gene expression microarray data, we observed a trend of increased FGFR2 expression in the rare homozygotes. This trend was confirmed using real-time (RT) PCR, with the difference between the rare and the common homozygotes yielding a Wilcox p-value of 0.028. To elucidate which SNPs might be responsible for this difference, we examined protein-DNA interactions for the eight most strongly disease-associated SNPs in different breast cell lines. We identify two cis-regulatory SNPs that alter binding affinity for transcription factors Oct-1/Runx2 and C/EBPbeta, and we demonstrate that both sites are occupied in vivo. In transient transfection experiments, the two SNPs can synergize giving rise to increased FGFR2 expression. We propose a model in which the Oct-1/Runx2 and C/EBPbeta binding sites in the disease-associated allele are able to lead to an increase in FGFR2 gene expression, thereby increasing the propensity for tumour formation.
Conflict of interest statement
Figures

References
-
- Grose R, Dickson C. Fibroblast growth factor signaling in tumorigenesis. Cytokine Growth Factor Rev. 2005;16:179–86. - PubMed
-
- Theodorou V, Kimm MA, Boer M, Wessels L, Theelen W, et al. MMTV insertional mutagenesis identifies genes, gene families and pathways involved in mammary cancer. Nat Genet. 2007;39:759–69. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

