Crystal structure of NFAT bound to the HIV-1 LTR tandem kappaB enhancer element
- PMID: 18462673
- PMCID: PMC2697820
- DOI: 10.1016/j.str.2008.01.020
Crystal structure of NFAT bound to the HIV-1 LTR tandem kappaB enhancer element
Abstract
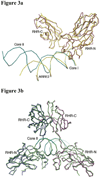
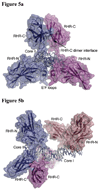
The host factor, nuclear factor of activated T-cells (NFAT), regulates the transcription and replication of HIV-1. Here, we have determined the crystal structure of the DNA binding domain of NFAT bound to the HIV-1 long terminal repeat (LTR) tandem kappaB enhancer element at 3.05 A resolution. NFAT binds as a dimer to the upstream kappaB site (Core II), but as a monomer to the 3' end of the downstream kappaB site (Core I). The DNA shows a significant bend near the 5' end of Core I, where a lysine residue from NFAT bound to the 3' end of Core II inserts into the minor groove and seems to cause DNA bases to flip out. Consistent with this structural feature, the 5' end of Core I become hypersensitive to dimethylsulfate in the in vivo footprinting upon transcriptional activation of the HIV-1 LTR. Our studies provide a basis for further investigating the functional mechanisms of NFAT in HIV-1 transcription and replication.
Figures







References
-
- Alcami J, Lain de Lera T, Folgueira L, Pedraza MA, Jacque JM, Bachelerie F, Noriega AR, Hay RT, Harrich D, Gaynor RB, et al. Absolute dependence on kappa B responsive elements for initiation and Tat-mediated amplification of HIV transcription in blood CD4 T lymphocytes. The EMBO journal. 1995;14:1552–1560. - PMC - PubMed
-
- Argyropoulos C, Mouzaki A. Immunosuppressive drugs in HIV disease. Current topics in medicinal chemistry. 2006;6:1769–1789. - PubMed
-
- Bounou S, Dumais N, Tremblay MJ. Attachment of human immunodeficiency virus-1 (HIV-1) particles bearing host-encoded B7-2 proteins leads to nuclear factor-kappa B-and nuclear factor of activated T cells-dependent activation of HIV-1 long terminal repeat transcription. The Journal of biological chemistry. 2001;276:6359–6369. - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta crystallographica. 1998;54:905–921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

