Global transcription in pluripotent embryonic stem cells
- PMID: 18462694
- PMCID: PMC2435228
- DOI: 10.1016/j.stem.2008.03.021
Global transcription in pluripotent embryonic stem cells
Abstract
The molecular mechanisms underlying pluripotency and lineage specification from embryonic stem cells (ESCs) are largely unclear. Differentiation pathways may be determined by the targeted activation of lineage-specific genes or by selective silencing of genome regions. Here we show that the ESC genome is transcriptionally globally hyperactive and undergoes large-scale silencing as cells differentiate. Normally silent repeat regions are active in ESCs, and tissue-specific genes are sporadically expressed at low levels. Whole-genome tiling arrays demonstrate widespread transcription in coding and noncoding regions in ESCs, whereas the transcriptional landscape becomes more discrete as differentiation proceeds. The transcriptional hyperactivity in ESCs is accompanied by disproportionate expression of chromatin-remodeling genes and the general transcription machinery. We propose that global transcription is a hallmark of pluripotent ESCs, contributing to their plasticity, and that lineage specification is driven by reduction of the transcribed portion of the genome.
Figures
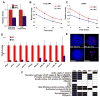
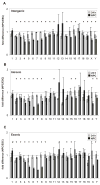

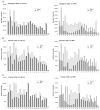
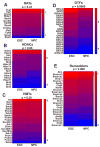

Comment in
-
Open chromatin and hypertranscription in embryonic stem cells.Cell Stem Cell. 2008 May 8;2(5):408-10. doi: 10.1016/j.stem.2008.04.006. Cell Stem Cell. 2008. PMID: 18462689
References
-
- Aoto T, Saitoh N, Ichimura T, Niwa H, Nakao M. Nuclear and chromatin reorganization in the MHC-Oct3/4 locus at developmental phases of embryonic stem cell differentiation. Dev Biol. 2006;298:354–367. - PubMed
-
- Araki R, Fukumura R, Sasaki N, Kasama Y, Suzuki N, Takahashi H, Tabata Y, Saito T, Abe M. More than 40,000 transcripts, including novel and noncoding transcripts, in mouse embryonic stem cells. Stem Cells. 2006;24:2522–2528. - PubMed
-
- Arney KL, Fisher AG. Epigenetic aspects of differentiation. J Cell Sci. 2004;117:4355–4363. - PubMed
-
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006a;8:532–538. - PubMed
-
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 2006b - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

