Loss of unc45a precipitates arteriovenous shunting in the aortic arches
- PMID: 18462713
- PMCID: PMC2483962
- DOI: 10.1016/j.ydbio.2008.03.022
Loss of unc45a precipitates arteriovenous shunting in the aortic arches
Abstract
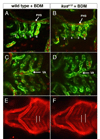
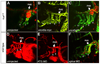
Aortic arch malformations are common congenital disorders that are frequently of unknown etiology. To gain insight into the factors that guide branchial aortic arch development, we examined the process by which these vessels assemble in wild type zebrafish embryos and in kurzschluss(tr12) (kus(tr12)) mutants. In wild type embryos, each branchial aortic arch first appears as an island of angioblasts in the lateral pharyngeal mesoderm, then elaborates by angiogenesis to connect to the lateral dorsal aorta and ventral aorta. In kus(tr12) mutants, angioblast formation and initial sprouting are normal, but aortic arches 5 and 6 fail to form a lumenized connection to the lateral dorsal aorta. Blood enters these blind-ending vessels from the ventral aorta, distending the arteries and precipitating fusion with an adjacent vein. This arteriovenous malformation (AVM), which shunts nearly all blood directly back to the heart, is not exclusively genetically programmed, as its formation correlates with blood flow and aortic arch enlargement. By positional cloning, we have identified a nonsense mutation in unc45a in kus(tr12) mutants. Our results are the first to ascribe a role for Unc45a, a putative myosin chaperone, in vertebrate development, and identify a novel mechanism by which an AVM can form.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

