The deep archaeal roots of eukaryotes
- PMID: 18463089
- PMCID: PMC2464739
- DOI: 10.1093/molbev/msn108
The deep archaeal roots of eukaryotes
Abstract
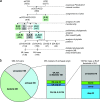
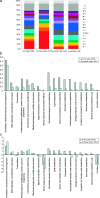
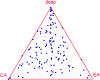
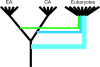
The set of conserved eukaryotic protein-coding genes includes distinct subsets one of which appears to be most closely related to and, by inference, derived from archaea, whereas another one appears to be of bacterial, possibly, endosymbiotic origin. The "archaeal" genes of eukaryotes, primarily, encode components of information-processing systems, whereas the "bacterial" genes are predominantly operational. The precise nature of the archaeo-eukaryotic relationship remains uncertain, and it has been variously argued that eukaryotic informational genes evolved from the homologous genes of Euryarchaeota or Crenarchaeota (the major branches of extant archaea) or that the origin of eukaryotes lies outside the known diversity of archaea. We describe a comprehensive set of 355 eukaryotic genes of apparent archaeal origin identified through ortholog detection and phylogenetic analysis. Phylogenetic hypothesis testing using constrained trees, combined with a systematic search for shared derived characters in the form of homologous inserts in conserved proteins, indicate that, for the majority of these genes, the preferred tree topology is one with the eukaryotic branch placed outside the extant diversity of archaea although small subsets of genes show crenarchaeal and euryarchaeal affinities. Thus, the archaeal genes in eukaryotes appear to descend from a distinct, ancient, and otherwise uncharacterized archaeal lineage that acquired some euryarchaeal and crenarchaeal genes via early horizontal gene transfer.
Figures

Similar articles
-
Comparative genomics of the Archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell.Genome Res. 1999 Jul;9(7):608-28. Genome Res. 1999. PMID: 10413400
-
Genome trees constructed using five different approaches suggest new major bacterial clades.BMC Evol Biol. 2001 Oct 20;1:8. doi: 10.1186/1471-2148-1-8. BMC Evol Biol. 2001. PMID: 11734060 Free PMC article.
-
The two-domain tree of life is linked to a new root for the Archaea.Proc Natl Acad Sci U S A. 2015 May 26;112(21):6670-5. doi: 10.1073/pnas.1420858112. Epub 2015 May 11. Proc Natl Acad Sci U S A. 2015. PMID: 25964353 Free PMC article.
-
Evolution of replicative DNA polymerases in archaea and their contributions to the eukaryotic replication machinery.Front Microbiol. 2014 Jul 21;5:354. doi: 10.3389/fmicb.2014.00354. eCollection 2014. Front Microbiol. 2014. PMID: 25101062 Free PMC article. Review.
-
The dispersed archaeal eukaryome and the complex archaeal ancestor of eukaryotes.Cold Spring Harb Perspect Biol. 2014 Apr 1;6(4):a016188. doi: 10.1101/cshperspect.a016188. Cold Spring Harb Perspect Biol. 2014. PMID: 24691961 Free PMC article. Review.
Cited by
-
The nature of the last universal common ancestor and its impact on the early Earth system.Nat Ecol Evol. 2024 Sep;8(9):1654-1666. doi: 10.1038/s41559-024-02461-1. Epub 2024 Jul 12. Nat Ecol Evol. 2024. PMID: 38997462 Free PMC article.
-
Hidden evolutionary complexity of Nucleo-Cytoplasmic Large DNA viruses of eukaryotes.Virol J. 2012 Aug 14;9:161. doi: 10.1186/1743-422X-9-161. Virol J. 2012. PMID: 22891861 Free PMC article.
-
Breaking through a phylogenetic impasse: a pair of associated archaea might have played host in the endosymbiotic origin of eukaryotes.Cell Biosci. 2012 Aug 22;2(1):29. doi: 10.1186/2045-3701-2-29. Cell Biosci. 2012. PMID: 22913376 Free PMC article.
-
Evolution: like any other science it is predictable.Philos Trans R Soc Lond B Biol Sci. 2010 Jan 12;365(1537):133-45. doi: 10.1098/rstb.2009.0154. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20008391 Free PMC article.
-
A New Family of DNA Viruses Causing Disease in Crustaceans from Diverse Aquatic Biomes.mBio. 2020 Jan 14;11(1):e02938-19. doi: 10.1128/mBio.02938-19. mBio. 2020. PMID: 31937645 Free PMC article.
References
-
- Anderson FE, Swofford DL. Should we be worried about long-branch attraction in real data sets? Investigations using metazoan 18S rDNA. Mol Phylogenet Evol. 2004;33:440–451. - PubMed
-
- Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. - PubMed
-
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

