Phylogenetic backgrounds and virulence profiles of atypical enteropathogenic Escherichia coli strains from a case-control study using multilocus sequence typing and DNA microarray analysis
- PMID: 18463209
- PMCID: PMC2446914
- DOI: 10.1128/JCM.01752-07
Phylogenetic backgrounds and virulence profiles of atypical enteropathogenic Escherichia coli strains from a case-control study using multilocus sequence typing and DNA microarray analysis
Abstract
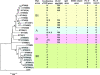
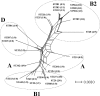
Atypical enteropathogenetic Escherichia coli (EPEC) strains are frequently detected in children with diarrhea but are also a common finding in healthy children. The aim of this study was to compare the phylogenetic ancestry and virulence characteristics of atypical (eae positive, stx and bfpA negative) EPEC strains from Norwegian children with (n = 37) or without (n = 19) diarrhea and to search for an association between phylogenetic ancestry and diarrhea. The strains were classified in phylogenetic groups by phylogenetic marker genes and in sequence types (STs) by multilocus sequence typing. Phylogenetic ancestry was compared to virulence characteristics based on DNA microarray analysis. Serotyping and pulsed-field gel electrophoresis (PFGE) were also performed. All four phylogenetic groups, 26 different STs, and 20 different clonal groups were represented among the 56 atypical EPEC strains. The strains were separated into three clusters by overall virulence gene profile; one large cluster with A, B1, and D strains and two clusters with group B2 strains. There was considerable heterogeneity in the PFGE profiles and serotypes, and almost half of the strains were O nontypeable. The efa1/lifA gene, previously shown to be statistically linked with diarrhea in this strain collection (J. E. Afset et al., J. Clin. Microbiol. 44:3703-3711, 2006), was present in 8 of 26 STs. The two phylogenetic groups B1 and D were weakly associated with diarrhea (P = 0.06 and P = 0.09, respectively). In contrast, group B2 was isolated most frequently from healthy controls (P = 0.05). In conclusion, the atypical EPEC strains were heterogeneous both phylogenetically and by virulence profile. Phylogenetic ancestry was less useful as a predictor of diarrhea than were specific virulence genes.
Figures


Similar articles
-
Distribution of non-locus of enterocyte effacement pathogenic island-related genes in Escherichia coli carrying eae from patients with diarrhea and healthy individuals in Japan.J Clin Microbiol. 2010 Nov;48(11):4107-14. doi: 10.1128/JCM.00677-10. Epub 2010 Sep 15. J Clin Microbiol. 2010. PMID: 20844211 Free PMC article.
-
Higher atypical enteropathogenic Escherichia coli (a-EPEC) bacterial loads in children with diarrhea are associated with PCR detection of the EHEC factor for adherence 1/lymphocyte inhibitory factor A (efa1/lifa) gene.Ann Clin Microbiol Antimicrob. 2017 Mar 23;16(1):16. doi: 10.1186/s12941-017-0188-y. Ann Clin Microbiol Antimicrob. 2017. PMID: 28330478 Free PMC article.
-
Virulence features of atypical enteropathogenic Escherichia coli identified by the eae(+) EAF-negative stx(-) genetic profile.Diagn Microbiol Infect Dis. 2009 Aug;64(4):357-65. doi: 10.1016/j.diagmicrobio.2009.03.025. Epub 2009 May 12. Diagn Microbiol Infect Dis. 2009. PMID: 19442475
-
Enteropathogenic Escherichia coli: foe or innocent bystander?Clin Microbiol Infect. 2015 Aug;21(8):729-34. doi: 10.1016/j.cmi.2015.01.015. Epub 2015 Jan 28. Clin Microbiol Infect. 2015. PMID: 25726041 Free PMC article. Review.
-
New insights into the epidemiology of enteropathogenic Escherichia coli infection.Trans R Soc Trop Med Hyg. 2008 Sep;102(9):852-6. doi: 10.1016/j.trstmh.2008.03.017. Epub 2008 May 2. Trans R Soc Trop Med Hyg. 2008. PMID: 18455741 Free PMC article. Review.
Cited by
-
Occurrence of potentially human-pathogenic Escherichia coli O103 in Norwegian sheep.Appl Environ Microbiol. 2013 Dec;79(23):7502-9. doi: 10.1128/AEM.01825-13. Epub 2013 Sep 27. Appl Environ Microbiol. 2013. PMID: 24077709 Free PMC article.
-
Molecular subtyping and distribution of the serine protease from shiga toxin-producing Escherichia coli among atypical enteropathogenic E. coli strains.Appl Environ Microbiol. 2009 Apr;75(7):2246-9. doi: 10.1128/AEM.01957-08. Epub 2009 Jan 9. Appl Environ Microbiol. 2009. PMID: 19139236 Free PMC article.
-
Association of nucleotide polymorphisms within the O-antigen gene cluster of Escherichia coli O26, O45, O103, O111, O121, and O145 with serogroups and genetic subtypes.Appl Environ Microbiol. 2012 Sep;78(18):6689-703. doi: 10.1128/AEM.01259-12. Epub 2012 Jul 13. Appl Environ Microbiol. 2012. PMID: 22798363 Free PMC article.
-
Phylogenetic classification of Escherichia coli O157:H7 strains of human and bovine origin using a novel set of nucleotide polymorphisms.Genome Biol. 2009;10(5):R56. doi: 10.1186/gb-2009-10-5-r56. Epub 2009 May 22. Genome Biol. 2009. PMID: 19463166 Free PMC article.
-
Comparisons of infant Escherichia coli isolates link genomic profiles with adaptation to the ecological niche.BMC Genomics. 2013 Feb 5;14:81. doi: 10.1186/1471-2164-14-81. BMC Genomics. 2013. PMID: 23384204 Free PMC article.
References
-
- Afset, J. E., L. Bevanger, P. Romundstad, and K. Bergh. 2004. Association of atypical enteropathogenic Escherichia coli (EPEC) with prolonged diarrhoea. J. Med. Microbiol. 531137-1144. - PubMed
-
- Aktan, I., K. A. Sprigings, R. M. La Ragione, L. M. Faulkner, G. A. Paiba, and M. J. Woodward. 2004. Characterisation of attaching-effacing Escherichia coli isolated from animals at slaughter in England and Wales. Vet. Microbiol. 10243-53. - PubMed
-
- Alikhani, M. Y., A. Mirsalehian, and M. M. Aslani. 2006. Detection of typical and atypical enteropathogenic Escherichia coli (EPEC) in Iranian children with and without diarrhoea. J. Med. Microbiol. 551159-1163. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

