Transcription induces strand-specific mutations at the 5' end of human genes
- PMID: 18463301
- PMCID: PMC2493432
- DOI: 10.1101/gr.076570.108
Transcription induces strand-specific mutations at the 5' end of human genes
Abstract
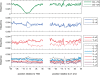
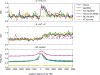
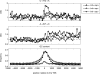
A regional analysis of nucleotide substitution rates along human genes and their flanking regions allows us to quantify the effect of mutational mechanisms associated with transcription in germ line cells. Our analysis reveals three distinct patterns of substitution rates. First, a sharp decline in the deamination rate of methylated CpG dinucleotides, which is observed in the vicinity of the 5' end of genes. Second, a strand asymmetry in complementary substitution rates, which extends from the 5' end to 1 kbp downstream from the 3' end, associated with transcription-coupled repair. Finally, a localized strand asymmetry, an excess of C-->T over G-->A substitution in the nontemplate strand confined to the first 1-2 kbp downstream of the 5' end of genes. We hypothesize that higher exposure of the nontemplate strand near the 5' end of genes leads to a higher cytosine deamination rate. Up to now, only the somatic hypermutation (SHM) pathway has been known to mediate localized and strand-specific mutagenic processes associated with transcription in mammalia. The mutational patterns in SHM are induced by cytosine deaminase, which just targets single-stranded DNA. This DNA conformation is induced by R-loops, which preferentially occur at the 5' ends of genes. We predict that R-loops are extensively formed in the beginning of transcribed regions in germ line cells.
Figures
References
-
- Aerts S., Thijs G., Dabrowski M., Moreau Y., De Moor B., Thijs G., Dabrowski M., Moreau Y., De Moor B., Dabrowski M., Moreau Y., De Moor B., Moreau Y., De Moor B., De Moor B. Comprehensive analysis of the base composition around the transcription start site in Metazoa. BMC Genomics. 2004;5:34. doi: 10.1186/1471-2164-5-34. - DOI - PMC - PubMed
-
- Aguilera A., Gomez-Gonzalez B., Gomez-Gonzalez B. Genome instability: A mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. - PubMed
-
- Aladjem M.I. Replication in context: Dynamic regulation of DNA replication patterns in metazoans. Nat. Rev. Genet. 2007;8:588–600. - PubMed
-
- Arndt P.F., Hwa T., Hwa T. Identification and measurement of neighbor-dependent nucleotide substitution processes. Bioinformatics. 2005;21:2322–2328. - PubMed
-
- Arndt P.F., Petrov D.A., Hwa T., Petrov D.A., Hwa T., Hwa T. Distinct changes of genomic biases in nucleotide substitution at the time of mammalian radiation. Mol. Biol. Evol. 2003;20:1887–1896. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
