Understanding NF-kappaB signaling via mathematical modeling
- PMID: 18463616
- PMCID: PMC2424295
- DOI: 10.1038/msb.2008.30
Understanding NF-kappaB signaling via mathematical modeling
Abstract
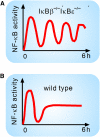
Mammalian inflammatory signaling, for which NF-kappaB is a principal transcription factor, is an exquisite example of how cellular signaling pathways can be regulated to produce different yet specific responses to different inflammatory insults. Mathematical models, tightly linked to experiment, have been instrumental in unraveling the forms of regulation in NF-kappaB signaling and their underlying molecular mechanisms. Our initial model of the IkappaB-NF-kappaB signaling module highlighted the role of negative feedback in the control of NF-kappaB temporal dynamics and gene expression. Subsequent studies sparked by this work have helped to characterize additional feedback loops, the input-output behavior of the module, crosstalk between multiple NF-kappaB-activating pathways, and NF-kappaB oscillations. We anticipate that computational techniques will enable further progress in the NF-kappaB field, and the signal transduction field in general, and we discuss potential upcoming developments.
Figures


References
-
- Beg AA, Sha WC, Bronson RT, Baltimore D (1995) Constitutive NF-kappa B activation, enhanced granulopoiesis, and neonatal lethality in I kappa B alpha-deficient mice. Genes Dev 9: 2736–2746 - PubMed
-
- Bren GD, Solan NJ, Miyoshi H, Pennington KN, Pobst LJ, Paya CV (2001) Transcription of the RelB gene is regulated by NF-kappaB. Oncogene 20: 7722–7733 - PubMed

