An endogenous small interfering RNA pathway in Drosophila
- PMID: 18463631
- PMCID: PMC2895258
- DOI: 10.1038/nature07007
An endogenous small interfering RNA pathway in Drosophila
Abstract
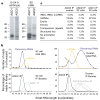
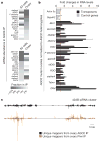
Drosophila endogenous small RNAs are categorized according to their mechanisms of biogenesis and the Argonaute protein to which they bind. MicroRNAs are a class of ubiquitously expressed RNAs of approximately 22 nucleotides in length, which arise from structured precursors through the action of Drosha-Pasha and Dicer-1-Loquacious complexes. These join Argonaute-1 to regulate gene expression. A second endogenous small RNA class, the Piwi-interacting RNAs, bind Piwi proteins and suppress transposons. Piwi-interacting RNAs are restricted to the gonad, and at least a subset of these arises by Piwi-catalysed cleavage of single-stranded RNAs. Here we show that Drosophila generates a third small RNA class, endogenous small interfering RNAs, in both gonadal and somatic tissues. Production of these RNAs requires Dicer-2, but a subset depends preferentially on Loquacious rather than the canonical Dicer-2 partner, R2D2 (ref. 14). Endogenous small interfering RNAs arise both from convergent transcription units and from structured genomic loci in a tissue-specific fashion. They predominantly join Argonaute-2 and have the capacity, as a class, to target both protein-coding genes and mobile elements. These observations expand the repertoire of small RNAs in Drosophila, adding a class that blurs distinctions based on known biogenesis mechanisms and functional roles.
Figures


Comment in
-
Genomics: protein fossils live on as RNA.Nature. 2008 Jun 5;453(7196):729-31. doi: 10.1038/453729a. Nature. 2008. PMID: 18528383 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

