Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918
- PMID: 18463694
- PMCID: PMC2262849
- DOI: 10.1371/journal.ppat.1000012
Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918
Abstract
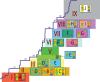
The H1N1 subtype of influenza A virus has caused substantial morbidity and mortality in humans, first documented in the global pandemic of 1918 and continuing to the present day. Despite this disease burden, the evolutionary history of the A/H1N1 virus is not well understood, particularly whether there is a virological basis for several notable epidemics of unusual severity in the 1940s and 1950s. Using a data set of 71 representative complete genome sequences sampled between 1918 and 2006, we show that segmental reassortment has played an important role in the genomic evolution of A/H1N1 since 1918. Specifically, we demonstrate that an A/H1N1 isolate from the 1947 epidemic acquired novel PB2 and HA genes through intra-subtype reassortment, which may explain the abrupt antigenic evolution of this virus. Similarly, the 1951 influenza epidemic may also have been associated with reassortant A/H1N1 viruses. Intra-subtype reassortment therefore appears to be a more important process in the evolution and epidemiology of H1N1 influenza A virus than previously realized.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






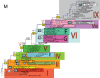

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

