Systematic analysis of pleiotropy in C. elegans early embryogenesis
- PMID: 18463698
- PMCID: PMC2265476
- DOI: 10.1371/journal.pcbi.1000003
Systematic analysis of pleiotropy in C. elegans early embryogenesis
Abstract
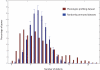
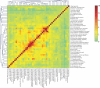
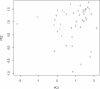
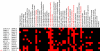
Pleiotropy refers to the phenomenon in which a single gene controls several distinct, and seemingly unrelated, phenotypic effects. We use C. elegans early embryogenesis as a model to conduct systematic studies of pleiotropy. We analyze high-throughput RNA interference (RNAi) data from C. elegans and identify "phenotypic signatures", which are sets of cellular defects indicative of certain biological functions. By matching phenotypic profiles to our identified signatures, we assign genes with complex phenotypic profiles to multiple functional classes. Overall, we observe that pleiotropy occurs extensively among genes involved in early embryogenesis, and a small proportion of these genes are highly pleiotropic. We hypothesize that genes involved in early embryogenesis are organized into partially overlapping functional modules, and that pleiotropic genes represent "connectors" between these modules. In support of this hypothesis, we find that highly pleiotropic genes tend to reside in central positions in protein-protein interaction networks, suggesting that pleiotropic genes act as connecting points between different protein complexes or pathways.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


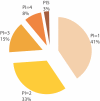
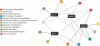
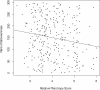
References
-
- Hartsfield JK, Jr, Hall BD, Grix AW, Kousseff BG, Salazar JF, et al. Pleiotropy in Coffin-Lowry syndrome: sensorineural hearing deficit and premature tooth loss as early manifestations. Am J Med Genet. 1993;45:552–557. - PubMed
-
- Foster KR, Shaulsky G, Strassmann JE, Queller DC, Thompson CR. Pleiotropy as a mechanism to stabilize cooperation. Nature. 2004;431:693–696. - PubMed
-
- Knight CG, Zitzmann N, Prabhakar S, Antrobus R, Dwek R, et al. Unraveling adaptive evolution: how a single point mutation affects the protein coregulation network. Nat Genet. 2006;38:1015–1022. - PubMed
-
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

