The signaling petri net-based simulator: a non-parametric strategy for characterizing the dynamics of cell-specific signaling networks
- PMID: 18463702
- PMCID: PMC2265486
- DOI: 10.1371/journal.pcbi.1000005
The signaling petri net-based simulator: a non-parametric strategy for characterizing the dynamics of cell-specific signaling networks
Abstract
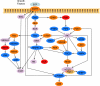
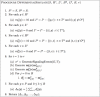
Reconstructing cellular signaling networks and understanding how they work are major endeavors in cell biology. The scale and complexity of these networks, however, render their analysis using experimental biology approaches alone very challenging. As a result, computational methods have been developed and combined with experimental biology approaches, producing powerful tools for the analysis of these networks. These computational methods mostly fall on either end of a spectrum of model parameterization. On one end is a class of structural network analysis methods; these typically use the network connectivity alone to generate hypotheses about global properties. On the other end is a class of dynamic network analysis methods; these use, in addition to the connectivity, kinetic parameters of the biochemical reactions to predict the network's dynamic behavior. These predictions provide detailed insights into the properties that determine aspects of the network's structure and behavior. However, the difficulty of obtaining numerical values of kinetic parameters is widely recognized to limit the applicability of this latter class of methods. Several researchers have observed that the connectivity of a network alone can provide significant insights into its dynamics. Motivated by this fundamental observation, we present the signaling Petri net, a non-parametric model of cellular signaling networks, and the signaling Petri net-based simulator, a Petri net execution strategy for characterizing the dynamics of signal flow through a signaling network using token distribution and sampling. The result is a very fast method, which can analyze large-scale networks, and provide insights into the trends of molecules' activity-levels in response to an external stimulus, based solely on the network's connectivity. We have implemented the signaling Petri net-based simulator in the PathwayOracle toolkit, which is publicly available at http://bioinfo.cs.rice.edu/pathwayoracle. Using this method, we studied a MAPK1,2 and AKT signaling network downstream from EGFR in two breast tumor cell lines. We analyzed, both experimentally and computationally, the activity level of several molecules in response to a targeted manipulation of TSC2 and mTOR-Raptor. The results from our method agreed with experimental results in greater than 90% of the cases considered, and in those where they did not agree, our approach provided valuable insights into discrepancies between known network connectivities and experimental observations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








References
-
- Hunter T. Signaling-2000 and beyond. Cell. 2000;100:113–127. - PubMed
-
- Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100:57–70. - PubMed
-
- Feldman DS, Carnes CA, Abraham WT, Bristow MR. Mechanisms of Disease: beta-adrenergic receptors alterations in signal transduction and pharmacogenomics in heart failure. Nature Clinical Practice Cardiovascular Medicine. 2005;2:475–483. - PubMed
-
- Belloni E, Muenke M, Roessier E, Traverse G, Siegel-Bartelt J, et al. Identification of Sonic hedgehog as a candidate gene responsible for holopro-sencephaly. Nat Genet. 1996;14:353–356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

