Mutations of different molecular origins exhibit contrasting patterns of regional substitution rate variation
- PMID: 18463707
- PMCID: PMC2265638
- DOI: 10.1371/journal.pcbi.1000015
Mutations of different molecular origins exhibit contrasting patterns of regional substitution rate variation
Abstract
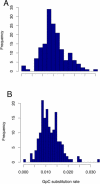
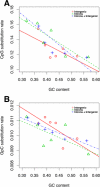
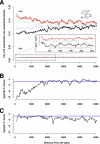
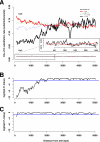
Transitions at CpG dinucleotides, referred to as "CpG substitutions", are a major mutational input into vertebrate genomes and a leading cause of human genetic disease. The prevalence of CpG substitutions is due to their mutational origin, which is dependent on DNA methylation. In comparison, other single nucleotide substitutions (for example those occurring at GpC dinucleotides) mainly arise from errors during DNA replication. Here we analyzed high quality BAC-based data from human, chimpanzee, and baboon to investigate regional variation of CpG substitution rates. We show that CpG substitutions occur approximately 15 times more frequently than other single nucleotide substitutions in primate genomes, and that they exhibit substantial regional variation. Patterns of CpG rate variation are consistent with differences in methylation level and susceptibility to subsequent deamination. In particular, we propose a "distance-decaying" hypothesis, positing that due to the molecular mechanism of a CpG substitution, rates are correlated with the stability of double-stranded DNA surrounding each CpG dinucleotide, and the effect of local DNA stability may decrease with distance from the CpG dinucleotide.Consistent with our "distance-decaying" hypothesis, rates of CpG substitution are strongly (negatively) correlated with regional G+C content. The influence of G+C content decays as the distance from the target CpG site increases. We estimate that the influence of local G+C content extends up to 1,500 approximately 2,000 bps centered on each CpG site. We also show that the distance-decaying relationship persisted when we controlled for the effect of long-range homogeneity of nucleotide composition. GpC sites, in contrast, do not exhibit such "distance-decaying" relationship. Our results highlight an example of the distinctive properties of methylation-dependent substitutions versus substitutions mostly arising from errors during DNA replication. Furthermore, the negative relationship between G+C content and CpG rates may provide an explanation for the observation that GC-rich SINEs show lower CpG rates than other repetitive elements.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
DNA structure constraint is probably a fundamental factor inducing CpG deficiency in bacteria.Bioinformatics. 2004 Dec 12;20(18):3336-45. doi: 10.1093/bioinformatics/bth393. Epub 2004 Jul 9. Bioinformatics. 2004. PMID: 15247101
-
Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes.Genome Res. 2010 Apr;20(4):447-57. doi: 10.1101/gr.098947.109. Epub 2010 Jan 26. Genome Res. 2010. PMID: 20103589 Free PMC article.
-
Identification and measurement of neighbor-dependent nucleotide substitution processes.Bioinformatics. 2005 May 15;21(10):2322-8. doi: 10.1093/bioinformatics/bti376. Epub 2005 Mar 15. Bioinformatics. 2005. PMID: 15769841
-
Monitoring methylation changes in cancer.Adv Biochem Eng Biotechnol. 2007;104:1-11. doi: 10.1007/10_024. Adv Biochem Eng Biotechnol. 2007. PMID: 17290816 Review.
-
The mutational spectrum of single base-pair substitutions causing human genetic disease: patterns and predictions.Hum Genet. 1990 Jun;85(1):55-74. doi: 10.1007/BF00276326. Hum Genet. 1990. PMID: 2192981 Review.
Cited by
-
The Avoidance of Purine Stretches by Cancer Mutations.Int J Mol Sci. 2024 Oct 15;25(20):11050. doi: 10.3390/ijms252011050. Int J Mol Sci. 2024. PMID: 39456831 Free PMC article.
-
DNA methylation and genome evolution in honeybee: gene length, expression, functional enrichment covary with the evolutionary signature of DNA methylation.Genome Biol Evol. 2010;2:770-80. doi: 10.1093/gbe/evq060. Epub 2010 Oct 5. Genome Biol Evol. 2010. PMID: 20924039 Free PMC article.
-
Substitution rate variation at human CpG sites correlates with non-CpG divergence, methylation level and GC content.Genome Biol. 2011 Jun 22;12(6):R58. doi: 10.1186/gb-2011-12-6-r58. Genome Biol. 2011. PMID: 21696599 Free PMC article.
-
Insights into Epigenome Evolution from Animal and Plant Methylomes.Genome Biol Evol. 2017 Nov 1;9(11):3189-3201. doi: 10.1093/gbe/evx203. Genome Biol Evol. 2017. PMID: 29036466 Free PMC article.
-
Mutation Rate Variation and Other Challenges in 2-LTR Dating of Primate Endogenous Retrovirus Integrations.J Mol Evol. 2025 Feb;93(1):62-82. doi: 10.1007/s00239-024-10225-5. Epub 2024 Dec 23. J Mol Evol. 2025. PMID: 39715846 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

