Genome landscapes and bacteriophage codon usage
- PMID: 18463708
- PMCID: PMC2266997
- DOI: 10.1371/journal.pcbi.1000001
Genome landscapes and bacteriophage codon usage
Abstract
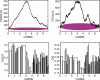
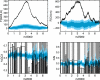
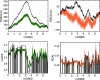
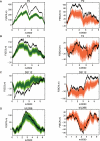
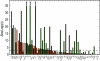
Across all kingdoms of biological life, protein-coding genes exhibit unequal usage of synonymous codons. Although alternative theories abound, translational selection has been accepted as an important mechanism that shapes the patterns of codon usage in prokaryotes and simple eukaryotes. Here we analyze patterns of codon usage across 74 diverse bacteriophages that infect E. coli, P. aeruginosa, and L. lactis as their primary host. We use the concept of a "genome landscape," which helps reveal non-trivial, long-range patterns in codon usage across a genome. We develop a series of randomization tests that allow us to interrogate the significance of one aspect of codon usage, such as GC content, while controlling for another aspect, such as adaptation to host-preferred codons. We find that 33 phage genomes exhibit highly non-random patterns in their GC3-content, use of host-preferred codons, or both. We show that the head and tail proteins of these phages exhibit significant bias towards host-preferred codons, relative to the non-structural phage proteins. Our results support the hypothesis of translational selection on viral genes for host-preferred codons, over a broad range of bacteriophages.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
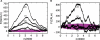

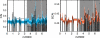
Similar articles
-
Genome dynamics, codon usage patterns and influencing factors in Aeromonas hydrophila phages.Virus Res. 2022 Oct 15;320:198900. doi: 10.1016/j.virusres.2022.198900. Epub 2022 Aug 24. Virus Res. 2022. PMID: 36029927
-
Significant differences in terms of codon usage bias between bacteriophage early and late genes: a comparative genomics analysis.BMC Genomics. 2017 Nov 13;18(1):866. doi: 10.1186/s12864-017-4248-7. BMC Genomics. 2017. PMID: 29132309 Free PMC article.
-
Differential codon adaptation between dsDNA and ssDNA phages in Escherichia coli.Mol Biol Evol. 2014 Jun;31(6):1606-17. doi: 10.1093/molbev/msu087. Epub 2014 Feb 27. Mol Biol Evol. 2014. PMID: 24586046 Free PMC article.
-
Codon usage bias: causative factors, quantification methods and genome-wide patterns: with emphasis on insect genomes.Biol Rev Camb Philos Soc. 2013 Feb;88(1):49-61. doi: 10.1111/j.1469-185X.2012.00242.x. Epub 2012 Aug 14. Biol Rev Camb Philos Soc. 2013. PMID: 22889422 Review.
-
DNA sequence evolution: the sounds of silence.Philos Trans R Soc Lond B Biol Sci. 1995 Sep 29;349(1329):241-7. doi: 10.1098/rstb.1995.0108. Philos Trans R Soc Lond B Biol Sci. 1995. PMID: 8577834 Review.
Cited by
-
CBDB: the codon bias database.BMC Bioinformatics. 2012 Apr 26;13:62. doi: 10.1186/1471-2105-13-62. BMC Bioinformatics. 2012. PMID: 22536831 Free PMC article.
-
Viral adaptation to host: a proteome-based analysis of codon usage and amino acid preferences.Mol Syst Biol. 2009;5:311. doi: 10.1038/msb.2009.71. Epub 2009 Oct 13. Mol Syst Biol. 2009. PMID: 19888206 Free PMC article.
-
Analysis of the codon usage pattern in Middle East Respiratory Syndrome Coronavirus.Oncotarget. 2017 Nov 27;8(66):110337-110349. doi: 10.18632/oncotarget.22738. eCollection 2017 Dec 15. Oncotarget. 2017. PMID: 29299151 Free PMC article.
-
Phage tRNAs evade tRNA-targeting host defenses through anticodon loop mutations.Elife. 2023 Jun 2;12:e85183. doi: 10.7554/eLife.85183. Elife. 2023. PMID: 37266569 Free PMC article.
-
Coevolution mechanisms that adapt viruses to genetic code variations implemented in their hosts.J Genet. 2016 Mar;95(1):3-12. doi: 10.1007/s12041-016-0612-7. J Genet. 2016. PMID: 27019427 No abstract available.
References
-
- Bernardi G. The human genome: Organization and evolutionary history. Annu Rev Genet. 1995;29:445–476. - PubMed
-
- Francino M, Ochman H. Isochores result from mutation not selection. Nature. 1999;400(6739):30–31. - PubMed
-
- Galtier N. Gene conversion drives gc content evolution in mammalian histones. Trends Genet. 2003;19:65–68. - PubMed
-
- Eyre-Walker A. An analysis of codon usage in mammals: selection or mutation bias? J Mol Evol. 1991;33:442–449. - PubMed
-
- Lawrence JG, Hartl DL. Unusual codon bias occurring within insertion sequences in Escherichia coli. Genetica. 1991;84:23–29. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

