Dynamic binding orientations direct activity of HIV reverse transcriptase
- PMID: 18464735
- PMCID: PMC2655135
- DOI: 10.1038/nature06941
Dynamic binding orientations direct activity of HIV reverse transcriptase
Abstract
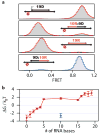
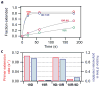
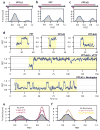
The reverse transcriptase of human immunodeficiency virus (HIV) catalyses a series of reactions to convert the single-stranded RNA genome of HIV into double-stranded DNA for host-cell integration. This task requires the reverse transcriptase to discriminate a variety of nucleic-acid substrates such that active sites of the enzyme are correctly positioned to support one of three catalytic functions: RNA-directed DNA synthesis, DNA-directed DNA synthesis and DNA-directed RNA hydrolysis. However, the mechanism by which substrates regulate reverse transcriptase activities remains unclear. Here we report distinct orientational dynamics of reverse transcriptase observed on different substrates with a single-molecule assay. The enzyme adopted opposite binding orientations on duplexes containing DNA or RNA primers, directing its DNA synthesis or RNA hydrolysis activity, respectively. On duplexes containing the unique polypurine RNA primers for plus-strand DNA synthesis, the enzyme can rapidly switch between the two orientations. The switching kinetics were regulated by cognate nucleotides and non-nucleoside reverse transcriptase inhibitors, a major class of anti-HIV drugs. These results indicate that the activities of reverse transcriptase are determined by its binding orientation on substrates.
Figures


Comment in
-
Molecular biology: an HIV secret uncovered.Nature. 2008 May 8;453(7192):169-70. doi: 10.1038/453169b. Nature. 2008. PMID: 18464731 No abstract available.
References
-
- Goff Stephen P. In: Fields Virology. Knipe DM, Howley PM, editors. Vol. 2. Lippincott Williams and Wilkins; Philadelphia: 2001. pp. 1871–1940.
-
- Champoux JJ. In: Reverse Transcriptase. Skalka AM, Goff SP, editors. Cold Spring Harbor Laboratory Press; New York: 1993. pp. 10–118.
-
- Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. - PubMed
-
- Baltimore D. Viral RNA-dependent DNA Polymerase: RNA-dependent DNA Polymerase in Virions of RNA Tumour Viruses. Nature. 1970;226:1209–1211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

