Suppression of plant resistance gene-based immunity by a fungal effector
- PMID: 18464895
- PMCID: PMC2330162
- DOI: 10.1371/journal.ppat.1000061
Suppression of plant resistance gene-based immunity by a fungal effector
Abstract
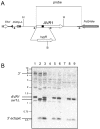
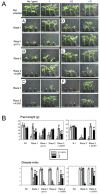
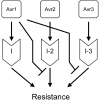
The innate immune system of plants consists of two layers. The first layer, called basal resistance, governs recognition of conserved microbial molecules and fends off most attempted invasions. The second layer is based on Resistance (R) genes that mediate recognition of effectors, proteins secreted by pathogens to suppress or evade basal resistance. Here, we show that a plant-pathogenic fungus secretes an effector that can both trigger and suppress R gene-based immunity. This effector, Avr1, is secreted by the xylem-invading fungus Fusarium oxysporum f.sp. lycopersici (Fol) and triggers disease resistance when the host plant, tomato, carries a matching R gene (I or I-1). At the same time, Avr1 suppresses the protective effect of two other R genes, I-2 and I-3. Based on these observations, we tentatively reconstruct the evolutionary arms race that has taken place between tomato R genes and effectors of Fol. This molecular analysis has revealed a hitherto unpredicted strategy for durable disease control based on resistance gene combinations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Stahl EA, Bishop JG. Plant-pathogen arms races at the molecular level. Curr Opin Plant Biol. 2000;3:299–304. - PubMed
-
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. - PubMed
-
- Catanzariti AM, Dodds PN, Ellis JG. Avirulence proteins from haustoria-forming pathogens. FEMS Microbiol Lett. 2007;269:181–188. - PubMed
-
- Rep M. Small proteins of plant-pathogenic fungi secreted during host colonization. FEMS Microbiol Lett. 2005;253:19–27. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

