Invasion of the Arabidopsis genome by the tobacco retrotransposon Tnt1 is controlled by reversible transcriptional gene silencing
- PMID: 18467467
- PMCID: PMC2442547
- DOI: 10.1104/pp.108.117846
Invasion of the Arabidopsis genome by the tobacco retrotransposon Tnt1 is controlled by reversible transcriptional gene silencing
Abstract
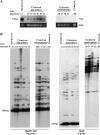
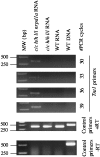
Long terminal repeat (LTR) retrotransposons are generally silent in plant genomes. However, they often constitute a large proportion of repeated sequences in plants. This suggests that their silencing is set up after a certain copy number is reached and/or that it can be released in some circumstances. We introduced the tobacco (Nicotiana tabacum) LTR retrotransposon Tnt1 into Arabidopsis (Arabidopsis thaliana), thus mimicking the horizontal transfer of a retrotransposon into a new host species and allowing us to study the regulatory mechanisms controlling its amplification. Tnt1 is transcriptionally silenced in Arabidopsis in a copy number-dependent manner. This silencing is associated with 24-nucleotide short-interfering RNAs targeting the promoter localized in the LTR region and with the non-CG site methylation of these sequences. Consequently, the silencing of Tnt1 is not released in methyltransferase1 mutants, in contrast to decrease in DNA methylation1 or polymerase IVa mutants. Stable reversion of Tnt1 silencing is obtained when the number of Tnt1 elements is reduced to two by genetic segregation. Our results support a model in which Tnt1 silencing in Arabidopsis occurs via an RNA-directed DNA methylation process. We further show that silencing can be partially overcome by some stresses.
Figures



Similar articles
-
The Tnt1 retrotransposon escapes silencing in tobacco, its natural host.PLoS One. 2012;7(3):e33816. doi: 10.1371/journal.pone.0033816. Epub 2012 Mar 30. PLoS One. 2012. PMID: 22479451 Free PMC article.
-
Expression of the tobacco Tnt1 retrotransposon promoter in heterologous species.Plant Mol Biol. 1994 Oct;26(1):393-402. doi: 10.1007/BF00039548. Plant Mol Biol. 1994. PMID: 7948885
-
Silencing of retrotransposons in arabidopsis and reactivation by the ddm1 mutation.Plant Cell. 2000 Mar;12(3):357-69. doi: 10.1105/tpc.12.3.357. Plant Cell. 2000. PMID: 10715322 Free PMC article.
-
The expression of the tobacco Tnt1 retrotransposon is linked to plant defense responses.Genetica. 1997;100(1-3):241-52. Genetica. 1997. PMID: 9440277 Review.
-
Stress activation and genomic impact of Tnt1 retrotransposons in Solanaceae.Cytogenet Genome Res. 2005;110(1-4):229-41. doi: 10.1159/000084957. Cytogenet Genome Res. 2005. PMID: 16093677 Review.
Cited by
-
Insertional mutagenesis using Tnt1 retrotransposon in potato.Plant Physiol. 2013 Sep;163(1):21-9. doi: 10.1104/pp.113.221903. Epub 2013 Jul 29. Plant Physiol. 2013. PMID: 23898040 Free PMC article.
-
MERE1, a low-copy-number copia-type retroelement in Medicago truncatula active during tissue culture.Plant Physiol. 2009 Nov;151(3):1250-63. doi: 10.1104/pp.109.138024. Epub 2009 Aug 5. Plant Physiol. 2009. PMID: 19656907 Free PMC article.
-
Reconstructing de novo silencing of an active plant retrotransposon.Nat Genet. 2013 Sep;45(9):1029-39. doi: 10.1038/ng.2703. Epub 2013 Jul 14. Nat Genet. 2013. PMID: 23852169
-
The stem cell state in plant development and in response to stress.Front Plant Sci. 2011 Oct 4;2:53. doi: 10.3389/fpls.2011.00053. eCollection 2011. Front Plant Sci. 2011. PMID: 22645540 Free PMC article.
-
Unravelling the Role of Epigenetic Modifications in Development and Reproduction of Angiosperms: A Critical Appraisal.Front Genet. 2022 May 18;13:819941. doi: 10.3389/fgene.2022.819941. eCollection 2022. Front Genet. 2022. PMID: 35664328 Free PMC article. Review.
References
-
- Andika IB, Kondo H, Rahim MD, Tamada T (2006) Lower levels of transgene silencing in roots is associated with reduced DNA methylation levels at non-symmetrical sites but not at symmetrical sites. Plant Mol Biol 60 423–435 - PubMed
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis. Nature 408 796–815 - PubMed
-
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82 259–266 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

