Hydrophile scanning as a complement to alanine scanning for exploring and manipulating protein-protein recognition: application to the Bim BH3 domain
- PMID: 18467496
- PMCID: PMC2442000
- DOI: 10.1110/ps.032896.107
Hydrophile scanning as a complement to alanine scanning for exploring and manipulating protein-protein recognition: application to the Bim BH3 domain
Abstract
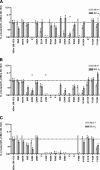
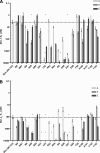
Alanine scanning has been widely employed as a method of identifying side chains that play important roles in protein-protein and protein-peptide interactions. Here we show how an analogous and complementary technique, hydrophile scanning, can provide additional insight on such interactions. Mutation of a wild-type residue to alanine removes most of the side-chain atoms, and the effect of this removal is typically interpreted to indicate contribution of the deleted side chain to the stability of the complex. Hydrophile scanning involves systematic mutation of wild-type residues to a cationic or anionic residue (lysine or glutamic acid, in this case). We find that the results of these mutations provide insights on interactions between polypeptide surfaces that are complementary to the information obtained via alanine scanning. We have applied this technique to a peptide that corresponds to the BH3 domain of the pro-apoptotic protein Bim. The wild-type Bim BH3 domain binds strongly to the anti-apoptotic proteins Bcl-x(L) and Mcl-1. Combining information from the alanine, lysine, and glutamic acid scans has enabled us to identify Bim BH3 domain mutants containing only two or three sequence changes that bind very selectively either to Bcl-x(L) or Mcl-1. Our findings suggest that hydrophile scanning may prove to be a broadly useful tool for revealing sources of protein-protein recognition and for engineering selectivity into natural sequences.
Figures

References
-
- Bruncko, M., Oost, T.K., Belli, B.A., Ding, H., Joseph, M.K., Kunzer, A., Martineau, D., McClellan, W.J., Mitten, M., Ng, S., et al. Studies leading to potent, dual inhibitors of bcl-2 and bcl-xL. J. Med. Chem. 2007;50:641–662. - PubMed
-
- Certo, M., Del Gaizo Moore, V., Nishino, M., Wei, G., Korsmeyer, S., Armstrong, S.A., Letai, A. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–365. - PubMed
-
- Chen, L., Willis, S.N., Wei, A., Smith, B.J., Fletcher, J.I., Hinds, M.G., Colman, P.M., Day, C.L., Adams, J.M., Huang, D.C.S. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell. 2005;17:393–403. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

