Viral IRES RNA structures and ribosome interactions
- PMID: 18468443
- PMCID: PMC2706518
- DOI: 10.1016/j.tibs.2008.04.007
Viral IRES RNA structures and ribosome interactions
Abstract
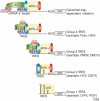
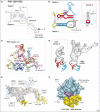
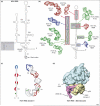
In eukaryotes, protein synthesis initiates primarily by a mechanism that requires a modified nucleotide 'cap' on the mRNA and also proteins that recruit and position the ribosome. Many pathogenic viruses use an alternative, cap-independent mechanism that substitutes RNA structure for the cap and many proteins. The RNAs driving this process are called internal ribosome-entry sites (IRESs) and some are able to bind the ribosome directly using a specific 3D RNA structure. Recent structures of IRES RNAs and IRES-ribosome complexes are revealing the structural basis of viral IRES' 'hijacking' of the protein-making machinery. It now seems that there are fundamental differences in the 3D structures used by different IRESs, although there are some common features in how they interact with ribosomes.
Figures


Similar articles
-
Viruses, IRESs, and a universal translation initiation mechanism.Biotechnol Genet Eng Rev. 2018 Apr;34(1):60-75. doi: 10.1080/02648725.2018.1471567. Epub 2018 May 28. Biotechnol Genet Eng Rev. 2018. PMID: 29804514 Review.
-
Structural basis for ribosome recruitment and manipulation by a viral IRES RNA.Science. 2006 Dec 1;314(5804):1450-4. doi: 10.1126/science.1133281. Epub 2006 Nov 23. Science. 2006. PMID: 17124290 Free PMC article.
-
Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome.Proc Natl Acad Sci U S A. 2011 Feb 1;108(5):1839-44. doi: 10.1073/pnas.1018582108. Epub 2011 Jan 18. Proc Natl Acad Sci U S A. 2011. PMID: 21245352 Free PMC article.
-
Structural and mechanistic insights into hepatitis C viral translation initiation.Nat Rev Microbiol. 2007 Jan;5(1):29-38. doi: 10.1038/nrmicro1558. Epub 2006 Nov 27. Nat Rev Microbiol. 2007. PMID: 17128284 Review.
-
A dynamic RNA loop in an IRES affects multiple steps of elongation factor-mediated translation initiation.Elife. 2015 Nov 2;4:e08146. doi: 10.7554/eLife.08146. Elife. 2015. PMID: 26523395 Free PMC article.
Cited by
-
Implications of RNG140 (caprin2)-mediated translational regulation in eye lens differentiation.J Biol Chem. 2020 Oct 30;295(44):15029-15044. doi: 10.1074/jbc.RA120.012715. Epub 2020 Aug 23. J Biol Chem. 2020. PMID: 32839273 Free PMC article.
-
Coronavirus takeover of host cell translation and intracellular antiviral response: a molecular perspective.EMBO J. 2024 Jan;43(2):151-167. doi: 10.1038/s44318-023-00019-8. Epub 2024 Jan 10. EMBO J. 2024. PMID: 38200146 Free PMC article. Review.
-
Production and Application of Multicistronic Constructs for Various Human Disease Therapies.Pharmaceutics. 2019 Nov 6;11(11):580. doi: 10.3390/pharmaceutics11110580. Pharmaceutics. 2019. PMID: 31698727 Free PMC article. Review.
-
Characterization of a canine homolog of hepatitis C virus.Proc Natl Acad Sci U S A. 2011 Jul 12;108(28):11608-13. doi: 10.1073/pnas.1101794108. Epub 2011 May 24. Proc Natl Acad Sci U S A. 2011. PMID: 21610165 Free PMC article.
-
Regulation of translation by ribosomal RNA pseudouridylation.Sci Adv. 2023 Aug 18;9(33):eadg8190. doi: 10.1126/sciadv.adg8190. Epub 2023 Aug 18. Sci Adv. 2023. PMID: 37595043 Free PMC article.
References
-
- Pestova T, et al. The mechanism of translation initiation in eukaryotes. In: Mathews MB, et al., editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; 2007. pp. 87–128.
-
- Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. - PubMed
-
- Doudna JA, Sarnow P. Translation initiation by viral internal ribosome entry sites. In: Mathews MB, et al., editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; 2007. pp. 129–153.
-
- Elroy-Stein O, Merrick WC. Translation initiation via cellular internal ribosome entry sites. In: Mathews MB, et al., editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; 2007. pp. 155–172.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

