Factors influencing local membrane curvature induction by N-BAR domains as revealed by molecular dynamics simulations
- PMID: 18469070
- PMCID: PMC2483763
- DOI: 10.1529/biophysj.107.121160
Factors influencing local membrane curvature induction by N-BAR domains as revealed by molecular dynamics simulations
Abstract
N-BAR domains are protein modules that bind to and induce curvature in membranes via a charged concave surface and N-terminal amphipathic helices. Recently, molecular dynamics simulations have demonstrated that the N-BAR domain can induce a strong local curvature that matches the curvature of the BAR domain surface facing the bilayer. Here we present further molecular dynamics simulations that examine in greater detail the roles of the concave surface and amphipathic helices in driving local membrane curvature. We find that the strong curvature induction observed in our previous simulations requires the stable presentation of the charged concave surface to the membrane and is not driven by the membrane-embedded amphipathic helices. Nevertheless, without these amphipathic helices embedded in the membrane, the N-BAR domain does not maintain a close association with the bilayer, and fails to drive membrane curvature. Increasing the membrane negative charge through the addition of PIP(2) facilitates closer association with the membrane in the absence of embedded helices. At sufficiently high concentrations, amphipathic helices embedded in the membrane drive membrane curvature independently of the BAR domain.
Figures
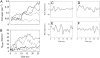
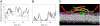



References
-
- McMahon, H. T., and J. L. Gallop. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 438:590–596. - PubMed
-
- Zimmerberg, J., and M. M. Kozlov. 2006. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 7:9–19. - PubMed
-
- McMahon, H. T., and I. G. Mills. 2004. COP and clathrin-coated vesicle budding: different pathways, common approaches. Curr. Opin. Cell Biol. 16:379–391. - PubMed
-
- Bi, X., R. A. Corpina, and J. Goldberg. 2002. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 419:271–277. - PubMed
-
- Nossal, R. 2001. Energetics of clathrin basket assembly. Traffic. 2:138–147. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

