DNA polymerases at the replication fork in eukaryotes
- PMID: 18471969
- PMCID: PMC2919282
- DOI: 10.1016/j.molcel.2008.04.011
DNA polymerases at the replication fork in eukaryotes
Abstract
The Kunkel laboratory has recently assigned polymerase (Pol) epsilon as the leading strand polymerase. In a recent issue of Molecular Cell, they now assign Pol delta as the lagging strand polymerase.
Figures
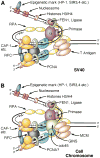
Comment on
-
Division of labor at the eukaryotic replication fork.Mol Cell. 2008 Apr 25;30(2):137-44. doi: 10.1016/j.molcel.2008.02.022. Mol Cell. 2008. PMID: 18439893 Free PMC article.
References
-
- Botchan M. Cell biology: a switch for S phase. Nature. 2007;445:272–274. - PubMed
-
- Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274:22283–22288. - PubMed
-
- Kesti T, Flick K, Keranen S, Syvaoja JE, Wittenberg C. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3:679–685. - PubMed
-
- Labib K, Gambus A. A key role for the GINS complex at DNA replication forks. Trends Cell Biol. 2007;17:271–278. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

