Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates
- PMID: 18471978
- PMCID: PMC2398651
- DOI: 10.1016/j.molcel.2008.02.028
Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates
Abstract
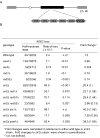
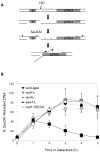
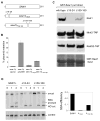
Elimination of a double-strand break (DSB) flanked by direct repeat sequences is mediated by single-strand annealing (SSA), which relies on a distinct set of gene products involving recombination, mismatch repair, and nucleotide excision repair. Here, we screened for yeast mutants defective in SSA with a plasmid-based SSA assay coupled to a barcode microarray readout. The screen identified Yal027Wp/Saw1 (single-strand annealing weakened 1) and Slx4 besides other known SSA proteins. Saw1 interacts physically with Rad1/Rad10, Msh2/Msh3, and Rad52 proteins, and cells lacking SLX4 or SAW1 accumulate recombination intermediates blocked at the Rad1/Rad10-dependent 3' flap cleavage step. Slx4 and Saw1 also contribute to the integrity of ribosomal DNA arrays. Saw1 mutants that fail to interact with Rad1, but retain interaction with Rad52 and Msh2, are defective in 3' flap removal and SSA repair. Deletion of SAW1 abolished association of Rad1 at SSA intermediates in vivo. We propose that Saw1 targets Rad1/Rad10 to Rad52-coated recombination intermediates.
Figures




References
-
- Bai Y, Symington LS. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. - PubMed
-
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

