Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy
- PMID: 18473723
- PMCID: PMC2443942
- DOI: 10.2174/187152008784220357
Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy
Abstract
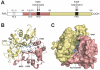
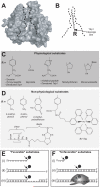
Tyrosyl-DNA phosphodiesterase 1 (Tdp1) is a recently discovered enzyme that catalyzes the hydrolysis of 3'-phosphotyrosyl bonds. Such linkages form in vivo following the DNA processing activity of topoisomerase I (Top1). For this reason, Tdp1 has been implicated in the repair of irreversible Top1-DNA covalent complexes, which can be generated by either exogenous or endogenous factors. Tdp1 has been regarded as a potential therapeutic co-target of Top1 in that it seemingly counteracts the effects of Top1 inhibitors, such as camptothecin and its clinically used derivatives. Thus, by reducing the repair of Top1-DNA lesions, Tdp1 inhibitors have the potential to augment the anticancer activity of Top1 inhibitors provided there is a presence of genetic abnormalities related to DNA checkpoint and repair pathways. Human Tdp1 can also hydrolyze other 3'-end DNA alterations including 3'-phosphoglycolates and 3'-abasic sites indicating it may function as a general 3'-DNA phosphodiesterase and repair enzyme. The importance of Tdp1 in humans is highlighted by the observation that a recessive mutation in the human TDP1 gene is responsible for the inherited disorder, spinocerebellar ataxia with axonal neuropathy (SCAN1). This review provides a summary of the biochemical and cellular processes performed by Tdp1 as well as the rationale behind the development of Tdp1 inhibitors for anticancer therapy.
Figures



References
-
- Champoux JJ. Annu. Rev. Biochem. 2001;70:369. - PubMed
-
- Keck JL, Berger JM. Nat. Struct. Biol. 1999;6:900. - PubMed
-
- Pommier Y, Pourquier P, Fan Y, Strumberg D. Biochim. Biophys. Acta. 1998;1400:83. - PubMed
-
- Wang JC. Nat. Rev. Mol. Cell Biol. 2002;3:430. - PubMed
-
- Hsiang YH, Hertzberg R, Hecht S, Liu LF. J. Biol. Chem. 1985;260:14873. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

