RNAi mechanisms and applications
- PMID: 18474035
- PMCID: PMC2727154
- DOI: 10.2144/000112792
RNAi mechanisms and applications
Abstract
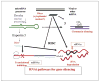
Within the past two decades we have become increasingly aware of the roles that RNAs play in regulation of gene expression. The RNA world was given a booster shot with the discovery of RNA interference (RNAi), a compendium of mechanisms involving small RNAs (less than 30 bases long) that regulate the expression of genes in a variety of eukaryotic organisms. Rapid progress in our understanding of RNAi-based mechanisms has led to applications of this powerful process in studies of gene function as well as in therapeutic applications for the treatment of disease. RNAi-based therapies involve two-dimensional drug designs using only identification of good Watson-Crick base pairing between the RNAi guide strand and the target, thereby resulting in rapid design and testing of RNAi triggers. To date there are several clinical trials using RNAi, and we should expect the list of new applications to grow at a phenomenal rate. This article summarizes our current knowledge about the mechanisms and applications of RNAi.
Figures
References
-
- Almeida R, Allshire RC. RNA silencing and genome regulation. Trends Cell Biol. 2005;15:251–258. - PubMed
-
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. - PubMed
-
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. - PubMed
-
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
