Proteasomal degradation restricts the nuclear lifespan of AID
- PMID: 18474627
- PMCID: PMC2413033
- DOI: 10.1084/jem.20070950
Proteasomal degradation restricts the nuclear lifespan of AID
Abstract
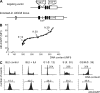
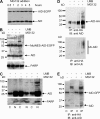
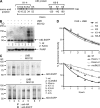
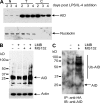
Activation-induced cytidine deaminase (AID) initiates all postrearrangement processes that diversify the immunoglobulin repertoire by specific deamination of cytidines at the immunoglobulin (Ig) locus. As uncontrolled expression of AID is potentially mutagenic, different types of regulation, particularly nucleocytoplasmic shuttling, restrict the likelihood of AID-deoxyribonucleic acid encounters. We studied additional mechanisms of regulation affecting the stability of the AID protein. No modulation of protein accumulation according to the cell cycle was observed in a Burkitt's lymphoma cell line. In contrast, the half-life of AID was markedly reduced in the nucleus, and this destabilization was accompanied by a polyubiquitination that was revealed in the presence of proteasome inhibitors. The same compartment-specific degradation was observed in activated mouse B cells, and also in a non-B cell line. No specific lysine residues could be linked to this degradation, so it remains unclear whether polyubiquitination proceeds through several alternatives sites or through the protein N terminus. The nuclear-restricted form of AID displayed enhanced mutagenicity at both Ig and non-Ig loci, most notably at TP53, suggesting that modulation of nuclear AID content through proteasomal degradation may represent another level of control of AID activity.
Figures


References
-
- Jung, D., C. Giallourakis, R. Mostoslavsky, and F.W. Alt. 2006. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 24:541–570. - PubMed
-
- Li, Z., C.J. Woo, M.D. Iglesias-Ussel, D. Ronai, and M.D. Scharff. 2004. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 18:1–11. - PubMed
-
- Muramatsu, M., V.S. Sankaranand, S. Anant, M. Sugai, K. Kinoshita, N.O. Davidson, and T. Honjo. 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274:18470–18476. - PubMed
-
- Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. - PubMed
-
- Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Gennery, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 102:565–575. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

