The transcription factor Nfix is essential for normal brain development
- PMID: 18477394
- PMCID: PMC2414869
- DOI: 10.1186/1471-213X-8-52
The transcription factor Nfix is essential for normal brain development
Abstract
Background: The Nuclear Factor I (NFI) multi-gene family encodes site-specific transcription factors essential for the development of a number of organ systems. We showed previously that Nfia-deficient mice exhibit agenesis of the corpus callosum and other forebrain defects; Nfib-deficient mice have defects in lung maturation and show callosal agenesis and forebrain defects resembling those seen in Nfia-deficient animals, while Nfic-deficient mice have defects in tooth root formation. Recently the Nfix gene has been disrupted and these studies indicated that there were largely uncharacterized defects in brain and skeletal development in Nfix-deficient mice.
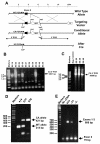
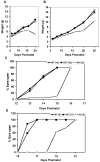
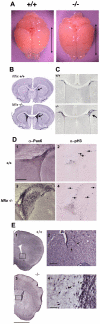
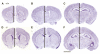
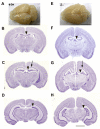
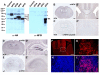
Results: Here we show that disruption of Nfix by Cre-recombinase mediated excision of the 2nd exon results in defects in brain development that differ from those seen in Nfia and Nfib KO mice. In particular, complete callosal agenesis is not seen in Nfix-/- mice but rather there appears to be an overabundance of aberrant Pax6- and doublecortin-positive cells in the lateral ventricles of Nfix-/- mice, increased brain weight, expansion of the cingulate cortex and entire brain along the dorsal ventral axis, and aberrant formation of the hippocampus. On standard lab chow Nfix-/- animals show a decreased growth rate from ~P8 to P14, lose weight from ~P14 to P22 and die at ~P22. If their food is supplemented with a soft dough chow from P10, Nfix-/- animals show a lag in weight gain from P8 to P20 but then increase their growth rate. A fraction of the animals survive to adulthood and are fertile. The weight loss correlates with delayed eye and ear canal opening and suggests a delay in the development of several epithelial structures in Nfix-/- animals.
Conclusion: These data show that Nfix is essential for normal brain development and may be required for neural stem cell homeostasis. The delays seen in eye and ear opening and the brain morphology defects appear independent of the nutritional deprivation, as rescue of perinatal lethality with soft dough does not eliminate these defects.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

