Dual KaiC-based oscillations constitute the circadian system of cyanobacteria
- PMID: 18477603
- PMCID: PMC2418587
- DOI: 10.1101/gad.1661808
Dual KaiC-based oscillations constitute the circadian system of cyanobacteria
Abstract
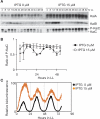
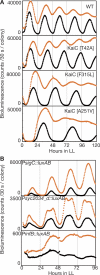
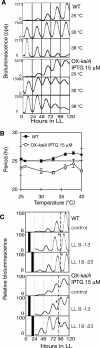
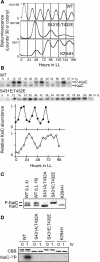
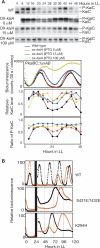
In the cyanobacterium Synechococcus elongatus PCC 7942, the KaiA, KaiB, and KaiC proteins are essential for the generation of circadian rhythms. Both in vivo and in vitro, phosphorylation of KaiC is regulated positively by KaiA and negatively by KaiB and shows circadian rhythmicity. The autonomous circadian cycling of KaiC phosphorylation is thought to be the basic pacemaker of the circadian clock and to control genome-wide gene expression in cyanobacteria. In this study, we found that temperature-compensated circadian oscillations of gene expression persisted even when KaiC was arrested in the phosphorylated state due to kaiA overexpression. Moreover, two phosphorylation mutants showed transcriptional oscillation with a long period. In kaiA-overexpressing and phosphorylation-deficient strains, KaiC oscillated and transient overexpression of phosphorylation-deficient kaiC reset the phase of the rhythm. These results suggest that transcription- and translation-based oscillations in KaiC abundance are also important for circadian rhythm generation in cyanobacteria. Furthermore, at low temperature, cyanobacteria can show circadian rhythms only when both the KaiC phosphorylation cycle and the transcription and translation cycle are intact. Our findings indicate that multiple coupled oscillatory systems based on the biochemical properties of KaiC are important to maintain robust and precise circadian rhythms in cyanobacteria.
Figures
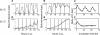
Comment in
-
Lego clocks: building a clock from parts.Genes Dev. 2008 Jun 1;22(11):1422-6. doi: 10.1101/gad.1686608. Genes Dev. 2008. PMID: 18519634 Free PMC article.
References
-
- Arita K., Hashimoto H., Igari K., Akaboshi M., Kutusna S., Sato M., Shimizu T. Structural and biochemical characterization of a cyanobacterium circadian clock-modifier protein. J. Biol. Chem. 2007;282:1128–1135. - PubMed
-
- Dunlap J.C., Loros J.J., DeCoursey P.J. Chronobiology: Biological timekeeping. Sinauer; Sunderland, MA: 2004.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
