Identification of a novel N-terminal hydrophobic sequence that targets proteins to lipid droplets
- PMID: 18477614
- PMCID: PMC2849272
- DOI: 10.1242/jcs.012013
Identification of a novel N-terminal hydrophobic sequence that targets proteins to lipid droplets
Abstract
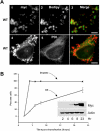
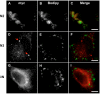
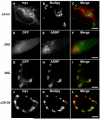
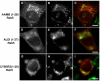
AAM-B is a putative methyltransferase that is a resident protein of lipid droplets. We have identified an N-terminal 28 amino acid hydrophobic sequence that is necessary and sufficient for targeting the protein to droplets. This sequence will also insert AAM-B into the endoplasmic reticulum (ER). A similar hydrophobic sequence (1-23) in the cytochrome p450 2C9 cannot substitute for 1-28 and only inserts AAM-B into the ER, which indicates that hydrophobicity and ER anchoring are not sufficient to reach the droplet. We found that a similar N-terminal hydrophobic sequence in cytochrome b5 reductase 3 and ALDI could also heterologously target proteins to droplets. Targeting is not affected by changing a conserved proline residue that potentially facilitates the formation of a hairpin loop to leucine. By contrast, targeting is blocked when AAM-B amino acids 59-64 or 65-70, situated downstream of the hydrophobic sequence, are changed to alanines. AAM-B-GFP expressed in Saccharomyces cerevisiae is also faithfully targeted to lipid bodies, indicating that the targeting mechanism is evolutionarily conserved. In conclusion, a class of hydrophobic sequences exists that when placed at the N-terminus of a protein will cause it to accumulate in droplets and in the ER.
Figures


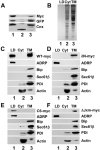
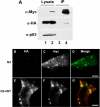
References
-
- Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, Zhao Y, Liu P. Dynamic activity of lipid droplets: protein phosphorylation and Gtp-mediated protein translocation. J. Proteome Res. 2007;6:3256–3265. - PubMed
-
- Blanchette-Mackie EJ, Dwyer NK, Barber T, Coxey RA, Takeda T, Rondinone CM, Theodorakis JL, Greenberg AS, Londos C. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J. Lipid Res. 1995;36:1211–1226. - PubMed
-
- Bracey MH, Cravatt BF, Stevens RC. Structural commonalities among integral membrane enzymes. FEBS Lett. 2004;567:159–165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

