Heterologous protein expression is enhanced by harmonizing the codon usage frequencies of the target gene with those of the expression host
- PMID: 18478103
- PMCID: PMC2364656
- DOI: 10.1371/journal.pone.0002189
Heterologous protein expression is enhanced by harmonizing the codon usage frequencies of the target gene with those of the expression host
Abstract
Synonymous codon replacement can change protein structure and function, indicating that protein structure depends on DNA sequence. During heterologous protein expression, low expression or formation of insoluble aggregates may be attributable to differences in synonymous codon usage between expression and natural hosts. This discordance may be particularly important during translation of the domain boundaries (link/end segments) that separate elements of higher ordered structure. Within such regions, ribosomal progression slows as the ribosome encounters clusters of infrequently used codons that preferentially encode a subset of amino acids. To replicate the modulation of such localized translation rates during heterologous expression, we used known relationships between codon usage frequencies and secondary protein structure to develop an algorithm ("codon harmonization") for identifying regions of slowly translated mRNA that are putatively associated with link/end segments. It then recommends synonymous replacement codons having usage frequencies in the heterologous expression host that are less than or equal to the usage frequencies of native codons in the native expression host. For protein regions other than these putative link/end segments, it recommends synonymous substitutions with codons having usage frequencies matched as nearly as possible to the native expression system. Previous application of this algorithm facilitated E. coli expression, manufacture and testing of two Plasmodium falciparum vaccine candidates. Here we describe the algorithm in detail and apply it to E. coli expression of three additional P. falciparum proteins. Expression of the "recoded" genes exceeded that of the native genes by 4- to 1,000-fold, representing levels suitable for vaccine manufacture. The proteins were soluble and reacted with a variety of functional conformation-specific mAbs suggesting that they were folded properly and had assumed native conformation. Codon harmonization may further provide a general strategy for improving the expression of soluble functional proteins during heterologous expression in hosts other than E. coli.
Conflict of interest statement
Figures


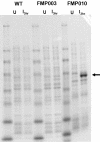
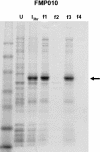
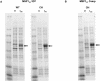
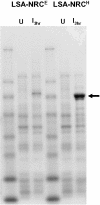
References
-
- Komar AA, Lesnik T, Reiss C. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 1999;462:387–391. - PubMed
-
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. - PubMed
-
- Adzhubei AA, Adzhubei IA, Krasheninnikov IA, Neidle S. Non-random usage of ‘degenerate’ codons is related to protein three-dimensional structure. FEBS Lett. 1996;399:78–82. - PubMed
-
- Kurland C, Gallant J. Errors of heterologous protein expression. Curr Opin Biotechnol. 1996;7:489–493. - PubMed
-
- Lindsley D, Gallant J, Guarneros G. Ribosome bypassing elicited by tRNA depletion. Mol Microbiol. 2003;48:1267–1274. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

