Highly conserved regimes of neighbor-base-dependent mutation generated the background primary-structural heterogeneities along vertebrate chromosomes
- PMID: 18478116
- PMCID: PMC2366069
- DOI: 10.1371/journal.pone.0002145
Highly conserved regimes of neighbor-base-dependent mutation generated the background primary-structural heterogeneities along vertebrate chromosomes
Erratum in
- PLoS One. 2013;8(9). doi:10.1371/annotation/bc789a4f-9fb7-41df-bc6f-d26203d09dbe
Abstract
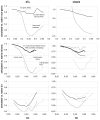
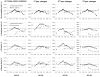
The content of guanine+cytosine varies markedly along the chromosomes of homeotherms and great effort has been devoted to studying this heterogeneity and its biological implications. Already before the DNA-sequencing era, however, it was established that the dinucleotides in the DNA of mammals in particular, and of most organisms in general, show striking over- and under-representations that cannot be explained by the base composition. Here we show that in the coding regions of vertebrates both GC content and codon occurrences are strongly correlated with such "motif preferences" even though we quantify the latter using an index that is not affected by the base composition, codon usage, and protein-sequence encoding. These correlations are likely to be the result of the long-term shaping of the primary structure of genic and non-genic DNA by a regime of mutation of which central features have been maintained by natural selection. We find indeed that these preferences are conserved in vertebrates even more rigidly than codon occurrences and we show that the occurrence-preference correlations are stronger in intronic and non-genic DNA, with the R(2)s reaching 99% when GC content is approximately 0.5. The mutation regime appears to be characterized by rates that depend markedly on the bases present at the site preceding and at that following each mutating site, because when we estimate such rates of neighbor-base-dependent mutation (NBDM) from substitutions retrieved from alignments of coding, intronic, and non-genic mammalian DNA sorted and grouped by GC content, they suffice to simulate DNA sequences in which motif occurrences and preferences as well as the correlations of motif preferences with GC content and with motif occurrences, are very similar to the mammalian ones. The best fit, however, is obtained with NBDM regimes lacking strand effects, which indicates that over the long term NBDM switches strands in the germline as one would expect for effects due to loosely contained background transcription. Finally, we show that human coding regions are less mutable under the estimated NBDM regimes than under matched context-independent mutation and that this entails marked differences between the spectra of amino-acid mutations that either mutation regime should generate. In the Discussion we examine the mechanisms likely to underlie NBDM heterogeneity along chromosomes and propose that it reflects how the diversity and activity of lesion-bypass polymerases (LBPs) track the landscapes of scheduled and non-scheduled genome repair, replication, and transcription during the cell cycle. We conclude that the primary structure of vertebrate genic DNA at and below the trinucleotide level has been governed over the long term by highly conserved regimes of NBDM which should be under direct natural selection because they alter drastically missense-mutation rates and hence the somatic and the germline mutational loads. Therefore, the non-coding DNA of vertebrates may have been shaped by NBDM only epiphenomenally, with non-genic DNA being affected mainly when found in the proximity of genes.
Conflict of interest statement
Figures

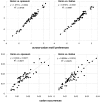




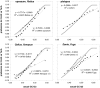











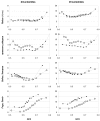

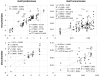









Similar articles
-
Mammalian mutation pressure, synonymous codon choice, and mRNA degradation.J Mol Evol. 2003 Dec;57(6):694-701. doi: 10.1007/s00239-003-2519-1. J Mol Evol. 2003. PMID: 14745538
-
A relationship between GC content and coding-sequence length.J Mol Evol. 1996 Sep;43(3):216-23. doi: 10.1007/BF02338829. J Mol Evol. 1996. PMID: 8703087
-
The evolution of transcription-associated biases of mutations across vertebrates.BMC Evol Biol. 2010 Jun 18;10:187. doi: 10.1186/1471-2148-10-187. BMC Evol Biol. 2010. PMID: 20565875 Free PMC article.
-
The vertebrate genome: isochores and evolution.Mol Biol Evol. 1993 Jan;10(1):186-204. doi: 10.1093/oxfordjournals.molbev.a039994. Mol Biol Evol. 1993. PMID: 8450755 Review.
-
Isochores and the evolutionary genomics of vertebrates.Gene. 2000 Jan 4;241(1):3-17. doi: 10.1016/s0378-1119(99)00485-0. Gene. 2000. PMID: 10607893 Review.
Cited by
-
You're one in a googol: optimizing genes for protein expression.J R Soc Interface. 2009 Aug 6;6 Suppl 4(Suppl 4):S467-76. doi: 10.1098/rsif.2008.0520.focus. Epub 2009 Mar 11. J R Soc Interface. 2009. PMID: 19324676 Free PMC article. Review.
-
Purifying selection in deeply conserved human enhancers is more consistent than in coding sequences.PLoS One. 2014 Jul 25;9(7):e103357. doi: 10.1371/journal.pone.0103357. eCollection 2014. PLoS One. 2014. PMID: 25062004 Free PMC article.
-
Evolution of hsp70 gene expression: a role for changes in AT-richness within promoters.PLoS One. 2011;6(5):e20308. doi: 10.1371/journal.pone.0020308. Epub 2011 May 31. PLoS One. 2011. PMID: 21655251 Free PMC article.
-
Was Wright right? The canonical genetic code is an empirical example of an adaptive peak in nature; deviant genetic codes evolved using adaptive bridges.J Mol Evol. 2010 Aug;71(2):87-99. doi: 10.1007/s00239-010-9373-8. Epub 2010 Aug 15. J Mol Evol. 2010. PMID: 20711776 Free PMC article. Review.
-
Introns form compositional clusters in parallel with the compositional clusters of the coding sequences to which they pertain.J Mol Evol. 2011 Jan;72(1):1-13. doi: 10.1007/s00239-010-9411-6. Epub 2010 Dec 4. J Mol Evol. 2011. PMID: 21132282
References
-
- Bernardi G. The compositional evolution of vertebrate genomes. Gene. 259((1–2)):31–43. - PubMed
-
- Bernardi G. Structural and Evolutionary Genomics - Natural Selection in Genome Evolution. Elsevier Science; 2005.
-
- King JL, Jukes TH. Non-Darwinian evolution. Science. 1969;164:788–798. - PubMed
-
- Antezana MA, Kreitman M. The nonrandom location of synonymous codons suggests that reading frame-independent forces have patterned codon preferences. Journal of Molecular Evolution. 1999;49((1)):36–43. - PubMed
-
- Nussinov R. Eukaryotic dinucleotide preference rules and their implications for degenerate codon usage. J Mol Biol. 1981a;149((1)):125–31. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

