Lack of aldose 1-epimerase in Hypocrea jecorina (anamorph Trichoderma reesei): a key to cellulase gene expression on lactose
- PMID: 18480250
- PMCID: PMC2438218
- DOI: 10.1073/pnas.0802789105
Lack of aldose 1-epimerase in Hypocrea jecorina (anamorph Trichoderma reesei): a key to cellulase gene expression on lactose
Abstract
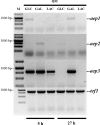
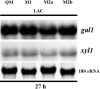
The heterodisaccharide lactose (1,4-O-beta-D-galactopyranosyl-D-glucose) induces cellulase formation in the ascomycete Hypocrea jecorina (= Trichoderma reesei). Lactose assimilation is slow, and the assimilation of its beta-D-galactose moiety depends mainly on the operation of a recently described reductive pathway and depends less on the Leloir pathway, which accepts only alpha-D-galactose. We therefore reasoned whether galactomutarotase [aldose 1-epimerase (AEP)] activity might limit lactose assimilation and thus influence cellulase formation. We identified three putative AEP-encoding genes (aep1, aep2, aep3) in H. jecorina, of which two encoded intracellular protein (AEP1 and AEP2) and one encoded an extracellular protein (AEP3). Although all three were transcribed, only the aep3 transcript was detected on lactose. However, no mutarotase activity was detected in the mycelia, their cell walls, or the extracellular medium during growth on lactose. Therefore, the effect of galactomutarotase activity on lactose assimilation was studied with H. jecorina strains expressing the C-terminal galactose mutarotase part of the Saccharomyces cerevisiae Gal10. These strains showed increased growth on lactose in a gene copy number-dependent manner, although their formation of extracellular beta-galactosidase activity and transcription of the genes encoding the first steps in the Leloir and the reductive pathway was similar to the parental strain QM9414. Cellulase gene transcription on lactose dramatically decreased in these strains, but remained unaffected during growth on cellulose. Our data show that cellulase induction in H. jecorina by lactose requires the beta-anomer of D-galactose and reveal the lack of mutarotase activity during growth on lactose as an important key for cellulase formation on this sugar.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
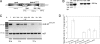



Similar articles
-
D-Galactose induces cellulase gene expression in Hypocrea jecorina at low growth rates.Microbiology (Reading). 2006 May;152(Pt 5):1507-1514. doi: 10.1099/mic.0.28719-0. Microbiology (Reading). 2006. PMID: 16622067
-
The galactokinase of Hypocrea jecorina is essential for cellulase induction by lactose but dispensable for growth on d-galactose.Mol Microbiol. 2004 Feb;51(4):1015-25. doi: 10.1046/j.1365-2958.2003.03901.x. Mol Microbiol. 2004. PMID: 14763977
-
Lactose metabolism and cellulase production in Hypocrea jecorina: the gal7 gene, encoding galactose-1-phosphate uridylyltransferase, is essential for growth on galactose but not for cellulase induction.Mol Genet Genomics. 2002 Mar;267(1):124-32. doi: 10.1007/s00438-002-0654-9. Epub 2002 Feb 22. Mol Genet Genomics. 2002. PMID: 11919723
-
Heterologous protein expression in Hypocrea jecorina: a historical perspective and new developments.Biotechnol Adv. 2015 Jan-Feb;33(1):142-154. doi: 10.1016/j.biotechadv.2014.11.009. Epub 2014 Dec 3. Biotechnol Adv. 2015. PMID: 25479282 Review.
-
[Transcriptional regulation of cellulases and hemicellulases gene in Hypocrea jecorina--a review].Wei Sheng Wu Xue Bao. 2010 Nov;50(11):1431-7. Wei Sheng Wu Xue Bao. 2010. PMID: 21268886 Review. Chinese.
Cited by
-
Comparative Phenotypic and Transcriptomic Analyses Provide Novel Insights into the Molecular Mechanism of Seed Germination in Response to Low Temperature Stress in Alfalfa.Int J Mol Sci. 2024 Jun 30;25(13):7244. doi: 10.3390/ijms25137244. Int J Mol Sci. 2024. PMID: 39000350 Free PMC article.
-
Differential protein expression profiling of Arabidopsis thaliana callus under microgravity on board the Chinese SZ-8 spacecraft.Planta. 2015 Feb;241(2):475-88. doi: 10.1007/s00425-014-2196-x. Epub 2014 Nov 6. Planta. 2015. PMID: 25374148
-
Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecorina.Biotechnol Biofuels. 2009 Sep 1;2:19. doi: 10.1186/1754-6834-2-19. Biotechnol Biofuels. 2009. PMID: 19723296 Free PMC article.
-
Intracellular Sugar Transporters Facilitate Cellulase Synthesis in Trichoderma reesei Using Lactose.Biomolecules. 2023 Feb 4;13(2):295. doi: 10.3390/biom13020295. Biomolecules. 2023. PMID: 36830663 Free PMC article.
-
Type Strains of Entomopathogenic Nematode-Symbiotic Bacterium Species, Xenorhabdus szentirmaii (EMC) and X. budapestensis (EMA), Are Exceptional Sources of Non-Ribosomal Templated, Large-Target-Spectral, Thermotolerant-Antimicrobial Peptides (by Both), and Iodinin (by EMC).Pathogens. 2022 Mar 11;11(3):342. doi: 10.3390/pathogens11030342. Pathogens. 2022. PMID: 35335666 Free PMC article. Review.
References
-
- Caputto R, Leloir LF, Cardini CE, Paladini AC. Isolation of the coenzyme of the galactose phosphate-glucose phosphate transformation. J Biol Chem. 1950;184:333–350. - PubMed
-
- Holden HM, Rayment I, Thoden JB. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J Biol Chem. 2003;278:43885–43888. - PubMed
-
- Bouffard GG, Rudd KE, Adhya SL. Dependence of lactose metabolism upon mutarotase encoded in the gal operon in Escherichia coli. J Mol Biol. 1994;244:269–278. - PubMed
-
- Wallenfels K, Hucho F, Herrmann K. Enzymatically catalyzed mutarotation of aldoses: Studies on aldose 1-epimerase from E. coli. Biochem Z. 1965;343:307–325. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

