Chaperone-dependent amyloid assembly protects cells from prion toxicity
- PMID: 18480252
- PMCID: PMC2438228
- DOI: 10.1073/pnas.0802593105
Chaperone-dependent amyloid assembly protects cells from prion toxicity
Abstract
Protein conformational diseases are associated with the aberrant accumulation of amyloid protein aggregates, but whether amyloid formation is cytotoxic or protective is unclear. To address this issue, we investigated a normally benign amyloid formed by the yeast prion [RNQ(+)]. Surprisingly, modest overexpression of Rnq1 protein was deadly, but only when preexisting Rnq1 was in the [RNQ(+)] prion conformation. Molecular chaperones protect against protein aggregation diseases and are generally believed to do so by solubilizing their substrates. The Hsp40 chaperone, Sis1, suppressed Rnq1 proteotoxicity, but instead of blocking Rnq1 protein aggregation, it stimulated conversion of soluble Rnq1 to [RNQ(+)] amyloid. Furthermore, interference with Sis1-mediated [RNQ(+)] amyloid formation exacerbated Rnq1 toxicity. These and other data establish that even subtle changes in the folding homeostasis of an amyloidogenic protein can create a severe proteotoxic gain-of-function phenotype and that chaperone-mediated amyloid assembly can be cytoprotective. The possible relevance of these findings to other phenomena, including prion-driven neurodegenerative diseases and heterokaryon incompatibility in fungi, is discussed.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
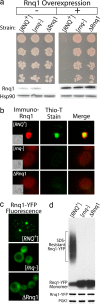


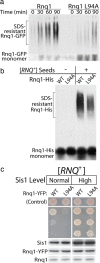
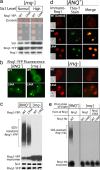
References
-
- Carrell RW, Lomas DA. Conformational disease. Lancet. 1997;350:134–138. - PubMed
-
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. - PubMed
-
- Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: Separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. - PubMed
-
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer's amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

