Prep1/Pbx2 complexes regulate CCL2 expression through the -2578 guanine polymorphism
- PMID: 18480829
- PMCID: PMC2570563
- DOI: 10.1038/gene.2008.33
Prep1/Pbx2 complexes regulate CCL2 expression through the -2578 guanine polymorphism
Abstract
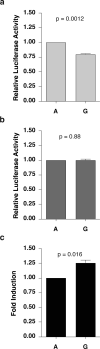
CC-chemokine ligand 2 (CCL2) is the major chemoattractant protein that recruits monocytes to sites of inflammation and increased expression of CCL2 is associated with numerous inflammatory diseases including human immunodeficiency virus-associated dementia (HIV-D). The -2578 guanine polymorphism in the CCL2 promoter has been associated with increased expression of CCL2 as well as pathogenesis of HIV-D; however, the molecular mechanism of regulation is unknown. We propose a molecular model for -2578 G-regulated CCL2 expression in astrocytes, which are major producers of CCL2 in the brain. The -2578 G polymorphism creates a consensus-binding site for the transcriptional regulator Prep1, which along with binding partner Pbx2, preferentially binds the -2578 G allele. CCL2 promoters harboring the G allele under unstimulated conditions exhibit a lower basal activity compared to the ancestral A allele. Upon interleukin-1 beta stimulation, Prep1/Pbx2 complexes maintain the ability to bind -2578 G alleles, yet transcription levels from promoters that harbor the A or G allele are equally activated, suggesting that the -2578 region does not influence CCL2 transcription under proinflammatory conditions. Therefore, promoters that harbor the -2578 G allele undergo a higher fold induction and by extension, individuals homozygous for -2578 G would be expected to exhibit hyper-responsive CCL2 phenotypes during periods of inflammation.
Figures
Comment in
-
Confirmation of differential binding of Interferon Regulatory Factor-1 (IRF-1) to the functional and HIV disease-influencing -2578 A/G polymorphism in CCL2.Genes Immun. 2009 Mar;10(2):197-8; author reply 199. doi: 10.1038/gene.2008.75. Epub 2008 Oct 16. Genes Immun. 2009. PMID: 18923432 No abstract available.
Similar articles
-
Regulation of CCL2 expression by an upstream TALE homeodomain protein-binding site that synergizes with the site created by the A-2578G SNP.PLoS One. 2011;6(7):e22052. doi: 10.1371/journal.pone.0022052. Epub 2011 Jul 8. PLoS One. 2011. PMID: 21760952 Free PMC article.
-
Confirmation of differential binding of Interferon Regulatory Factor-1 (IRF-1) to the functional and HIV disease-influencing -2578 A/G polymorphism in CCL2.Genes Immun. 2009 Mar;10(2):197-8; author reply 199. doi: 10.1038/gene.2008.75. Epub 2008 Oct 16. Genes Immun. 2009. PMID: 18923432 No abstract available.
-
The homeodomain Pbx2-Prep1 complex modulates hepatocyte nuclear factor 1alpha-mediated activation of the UDP-glucuronosyltransferase 2B17 gene.Mol Pharmacol. 2002 Jul;62(1):154-61. doi: 10.1124/mol.62.1.154. Mol Pharmacol. 2002. PMID: 12065766
-
The molecular structure and role of CCL2 (MCP-1) and C-C chemokine receptor CCR2 in skeletal biology and diseases.J Cell Physiol. 2021 Oct;236(10):7211-7222. doi: 10.1002/jcp.30375. Epub 2021 Mar 30. J Cell Physiol. 2021. PMID: 33782965 Review.
-
Monocyte chemoattractant protein-1 (MCP-1): an overview.J Interferon Cytokine Res. 2009 Jun;29(6):313-26. doi: 10.1089/jir.2008.0027. J Interferon Cytokine Res. 2009. PMID: 19441883 Free PMC article. Review.
Cited by
-
The rs1024611 regulatory region polymorphism is associated with CCL2 allelic expression imbalance.PLoS One. 2012;7(11):e49498. doi: 10.1371/journal.pone.0049498. Epub 2012 Nov 16. PLoS One. 2012. PMID: 23166687 Free PMC article.
-
Regulation of SIV mac 239 basal long terminal repeat activity and viral replication in macrophages: functional roles of two CCAAT/enhancer-binding protein beta sites in activation and interferon beta-mediated suppression.J Biol Chem. 2010 Jan 22;285(4):2258-73. doi: 10.1074/jbc.M109.075929. Epub 2009 Nov 20. J Biol Chem. 2010. PMID: 19933495 Free PMC article.
-
From genetic variants to therapeutic targets: insights into understanding rheumatoid arthritis.Front Immunol. 2025 Apr 1;16:1556971. doi: 10.3389/fimmu.2025.1556971. eCollection 2025. Front Immunol. 2025. PMID: 40236704 Free PMC article. Review.
-
CCL2/MCP-I genotype-phenotype relationship in latent tuberculosis infection.PLoS One. 2011;6(10):e25803. doi: 10.1371/journal.pone.0025803. Epub 2011 Oct 4. PLoS One. 2011. PMID: 21991356 Free PMC article.
-
Heme oxygenase-1 microsatellite polymorphism and haplotypes are associated with the development of acute respiratory distress syndrome.Intensive Care Med. 2009 Aug;35(8):1343-51. doi: 10.1007/s00134-009-1504-6. Epub 2009 Jun 13. Intensive Care Med. 2009. PMID: 19526221 Free PMC article.
References
-
- Leonard EJ, Yoshimura T. Human monocyte chemoattractant protein-1 (MCP-1). Immunol Today. 1990;11(3):97–101. - PubMed
-
- Fenoglio C, Galimberti D, Lovati C, Guidi I, Gatti A, Fogliarino S, et al. MCP-1 in Alzheimer's disease patients: A-2518G polymorphism and serum levels. Neurobiol Aging. 2004;25(9):1169–1173. - PubMed
-
- Craig MJ, Loberg RD. CCL2 (Monocyte Chemoattractant Protein-1) in cancer bone metastases. Cancer Metastasis Rev. 2006;25(4):611–619. - PubMed
-
- Galimberti D, Fenoglio C, Lovati C, Venturelli E, Guidi I, Corra B, et al. Serum MCP-1 levels are increased in mild cognitive impairment and mild Alzheimer's disease. Neurobiol Aging. 2006;27(12):1763–1768. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

