Isolation and characterization of novel microsatellite markers and their application for diversity assessment in cultivated groundnut (Arachis hypogaea)
- PMID: 18482440
- PMCID: PMC2416452
- DOI: 10.1186/1471-2229-8-55
Isolation and characterization of novel microsatellite markers and their application for diversity assessment in cultivated groundnut (Arachis hypogaea)
Abstract
Background: Cultivated peanut or groundnut (Arachis hypogaea L.) is the fourth most important oilseed crop in the world, grown mainly in tropical, subtropical and warm temperate climates. Due to its origin through a single and recent polyploidization event, followed by successive selection during breeding efforts, cultivated groundnut has a limited genetic background. In such species, microsatellite or simple sequence repeat (SSR) markers are very informative and useful for breeding applications. The low level of polymorphism in cultivated germplasm, however, warrants a need of larger number of polymorphic microsatellite markers for cultivated groundnut.
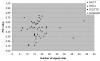
Results: A microsatellite-enriched library was constructed from the genotype TMV2. Sequencing of 720 putative SSR-positive clones from a total of 3,072 provided 490 SSRs. 71.2% of these SSRs were perfect type, 13.1% were imperfect and 15.7% were compound. Among these SSRs, the GT/CA repeat motifs were the most common (37.6%) followed by GA/CT repeat motifs (25.9%). The primer pairs could be designed for a total of 170 SSRs and were optimized initially on two genotypes. 104 (61.2%) primer pairs yielded scorable amplicon and 46 (44.2%) primers showed polymorphism among 32 cultivated groundnut genotypes. The polymorphic SSR markers detected 2 to 5 alleles with an average of 2.44 per locus. The polymorphic information content (PIC) value for these markers varied from 0.12 to 0.75 with an average of 0.46. Based on 112 alleles obtained by 46 markers, a phenogram was constructed to understand the relationships among the 32 genotypes. Majority of the genotypes representing subspecies hypogaea were grouped together in one cluster, while the genotypes belonging to subspecies fastigiata were grouped mainly under two clusters.
Conclusion: Newly developed set of 104 markers extends the repertoire of SSR markers for cultivated groundnut. These markers showed a good level of PIC value in cultivated germplasm and therefore would be very useful for germplasm analysis, linkage mapping, diversity studies and phylogenetic relationships in cultivated groundnut as well as related Arachis species.
Figures


References
-
- Dwivedi SL, Crouch JH. Proceedings of a Workshop for the Asian Development Bank supported project on molecular breeding of sorghum, groundnut and chickpea. ICRISAT. 2003. pp. 28–43.
-
- Weiss EA. Oilseed Crops. London, UK: Longman; 1983. p. 660.
-
- Savage GP, Keenan JI. The composition and nutritive value of groundnut kernels. In: Smart J, editor. The groundnut crop: A scientific basis for improvement. London, UK: Chapman and Hall; 1994. pp. 173–213.
-
- Stalker HT, Moss JP. Speciation, cytogenetics and utilization of Arachis species. Adv Agron. 1987;41:1–39.
-
- Young ND, Weeden NF, Kochert G. Genome mapping in legumes (Fam. Fabaceae) In: Paterson AH, editor. Genome Mapping in Plants. Austin, USA: Landes Company; 1996. pp. 211–227.
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

