Beyond tissueInfo: functional prediction using tissue expression profile similarity searches
- PMID: 18483083
- PMCID: PMC2441795
- DOI: 10.1093/nar/gkn233
Beyond tissueInfo: functional prediction using tissue expression profile similarity searches
Abstract
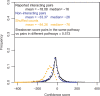
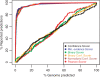
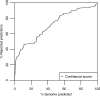
We present and validate tissue expression profile similarity searches (TEPSS), a computational approach to identify transcripts that share similar tissue expression profiles to one or more transcripts in a group of interest. We evaluated TEPSS for its ability to discriminate between pairs of transcripts coding for interacting proteins and non-interacting pairs. We found that ordering protein-protein pairs by TEPSS score produces sets significantly enriched in reported pairs of interacting proteins [interacting versus non-interacting pairs, Odds-ratio (OR) = 157.57, 95% confidence interval (CI) (36.81-375.51) at 1% coverage, employing a large dataset of about 50 000 human protein interactions]. When used with multiple transcripts as input, we find that TEPSS can predict non-obvious members of the cytosolic ribosome. We used TEPSS to predict S-nitrosylation (SNO) protein targets from a set of brain proteins that undergo SNO upon exposure to physiological levels of S-nitrosoglutathione in vitro. While some of the top TEPSS predictions have been validated independently, several of the strongest SNO TEPSS predictions await experimental validation. Our data indicate that TEPSS is an effective and flexible approach to functional prediction. Since the approach does not use sequence similarity, we expect that TEPSS will be useful for various gene discovery applications. TEPSS programs and data are distributed at http://icb.med.cornell.edu/crt/tepss/index.xml.
Figures

References
-
- Adams M, Kelley J, Gocayne J, Dubnick M, Polymeropoulos M, Xiao H, Merril C, Wu A, Olde B, Moreno R. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. - PubMed
-
- Boguski M, Lowe T, Tolstoshev C. dbEST—database for “expressed sequence tags”. Nat. Genet. 1993;4:332–333. - PubMed
-
- O’Dowd B, Nguyen T, Marchese A, Cheng R, Lynch K, Heng H, Kolakowski L, George S. Discovery of three novel G-protein-coupled receptor genes. Genomics. 1998;47:310–313. - PubMed
-
- Marchese A, Nguyen T, Malik P, Xu S, Cheng R, Xie Z, Heng H, George S, Kolakowski L, O’Dowd B. Cloning genes encoding receptors related to chemoattractant receptors. Genomics. 1998;50:281–286. - PubMed
-
- Haridas V, Ni J, Meager A, Su J, Yu G, Zhai Y, Kyaw H, Akama K, Hu J, Van Eldik L, et al. TRANK, a novel cytokine that activates NF-kappa B and c-jun N-terminal kinase. J. Immunol. 1998;161:1–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

